Final ID: 4125887
Aladyn Individual: Dynamic Individual Comorbidity Modeling for Genomic Discovery and Clinical Prediction
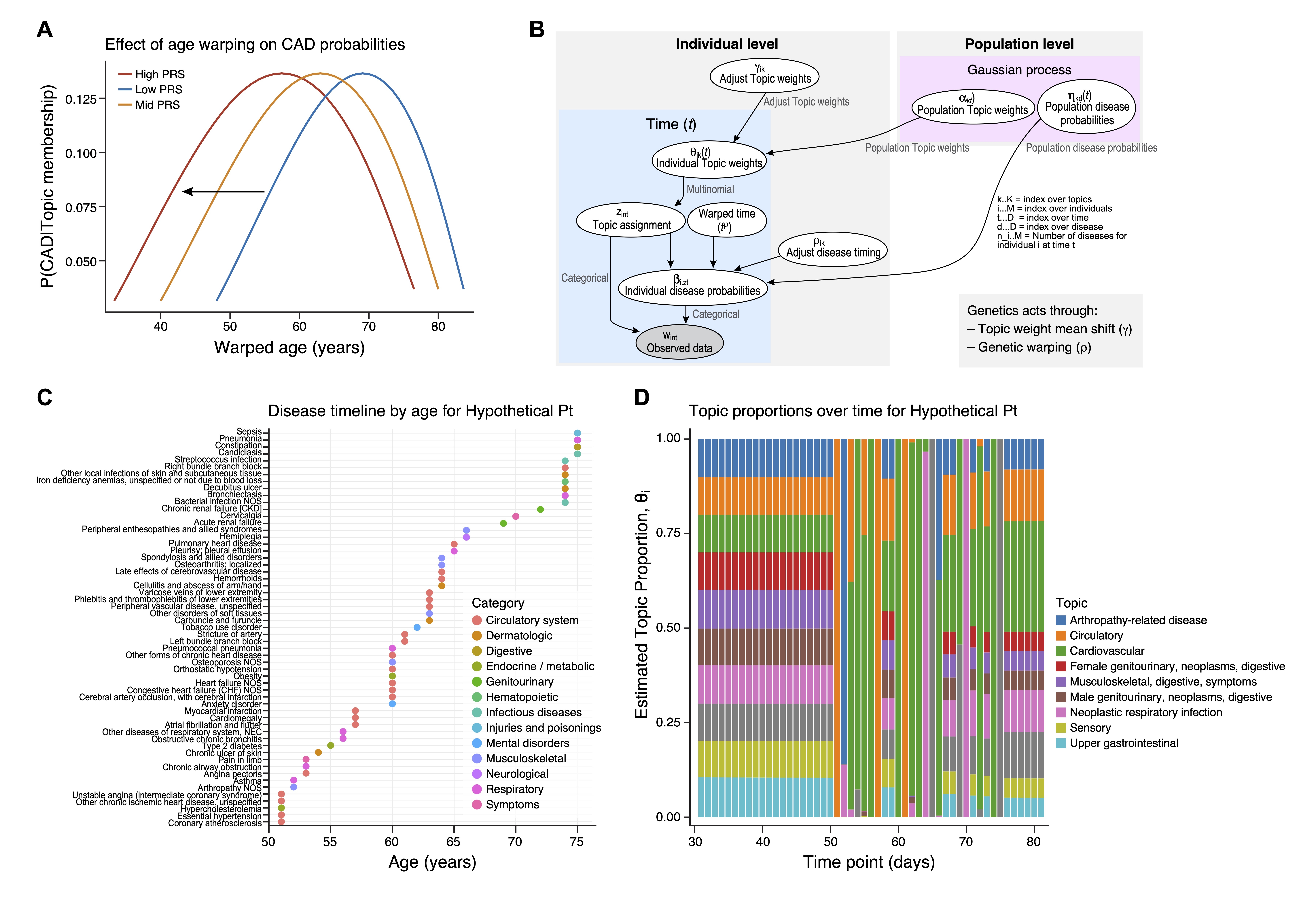
Abstract Body (Do not enter title and authors here): Intro: Considering comorbidity patterns for both discovery and prediction of individual disease trajectories is essential for personalized medicine. Existing methods often lack genetic integration and interpretability. We develop an approach suited for dynamic environments that combines genetics with individual and population level-trajectories for enhanced discovery and prediction. Method: Our method employs a dynamic Bayesian hierarchical latent allocation framework to learn and predict disease trajectories. Leveraging longitudinal electronic health care data from 485,698 UK Biobank participants and 287,012 All of Us Research Program participants, we aim to learn individual and population level disease trajectories for 352 distinct conditions. Genetic risks inform the model in two fundamental ways: First, we introduce a genetic warping parameter that recognizes the tendency of an individual to experience events earlier or later within these profiles while retaining the relative order of progression (1a). Second, we introduce a gaussian process to combine individual and population level trends in comorbidity profile prevalence over time (1b), whose mean is augmented by genetic predilection. Finally, we allow for profile weights to update with each new diagnoses, merging prediction and classification. Results: Our results indicate stark disparities in disease progression; individuals at high genetic risk for myocardial infarction experience events nearly a decade earlier than those at low risk (median age of onset 53.7 [53.1-53.9] vs 62.6 years [61.4-63.1]) (1b). Our model dynamically updates and predicts correct diagnoses 79.5% more frequently than existing topic-modeling approaches (1c,d). The dynamic approach adjusts disease profiles by at least 11.7% annually for 50% of individuals, with 65% experiencing over a 10% change in comorbidity profiles. Simulation results confirm the model's superior accuracy [51.4-41%], precision [50.9 vs 39.9%], and recall [48.4 vs 39.2%] compared to fixed-weight approaches. Summary: We offer a novel approach for integrating genetic risk with dynamic modeling of complex disease profiles for joint biologic discovery and personalized clinical prediction over the lifetime.
More abstracts on this topic:
A Heart-pounding Case of Cardiomyopathy in Pregnancy
Tran Linh, Everitt Ian, Vaught Arthur, Barth Andreas, Minhas Anum
A Qualitative Study of Perspectives on South Asian Dietary Practices: Exploring a Framework for Culturally Tailored Food-is-Medicine InterventionsKaloth Srivarsha, Fitzgerald Nurgul, Bacalia Karen Mae, Kalbag Aparna, Setoguchi Soko