Final ID: TAC238
Predicting Peptide-Human Leukocyte Antigen and Isolevuglandin Immunogenicity in Hypertension Using Structure-based Deep Learning
Abstract Body: Introduction
Isolevuglandin (IsoLG)-adducted peptides contribute to hypertension by enhancing class I human leukocyte antigen (HLA)-mediated T cell activation. The capacity of different HLA alleles to present IsoLG-modified peptides varies, influencing immune activation and disease progression. To elucidate the molecular determinants of peptide-HLA (pHLA) interactions, we combined cell-based peptide binding assays, structural modeling, and deep learning to predict pHLA binding affinity (IC50) and the presentation of IsoLG-modified epitopes.
Methods
IsoLG presentation was quantified in K562 cells, each transfected to express a single HLA (45 total, covering 90% of the US population), using flow cytometry-based Förster resonance energy transfer (FRET), which measures peptide-HLA proximity via donor-acceptor fluorescence. In computational studies, we filtered experimental pHLA binding data from the Immune Epitope Databank (IEDB) and modeled 49,268 structures using Rosetta and AlphaFold. Graph embeddings were constructed for pHLA complexes and unbound HLA alleles, incorporating per-residue and pairwise binding energies, and IC50 values. HLA embeddings were enriched with IsoLG FRET data to train a graph neural network, which combines information from both local molecular structure and broader interaction patterns using graph convolution and self-attention. The approach accommodates missing data and variable graph sizes using custom loss (error tracking) functions to manage outliers and incomplete measurements.
Results
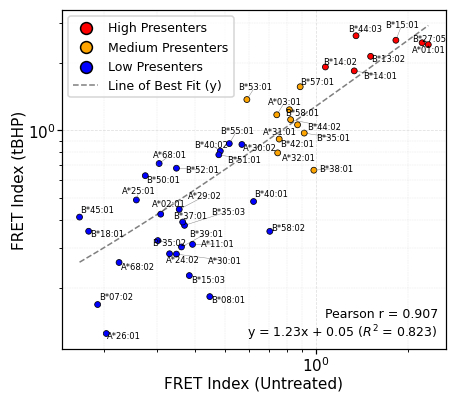
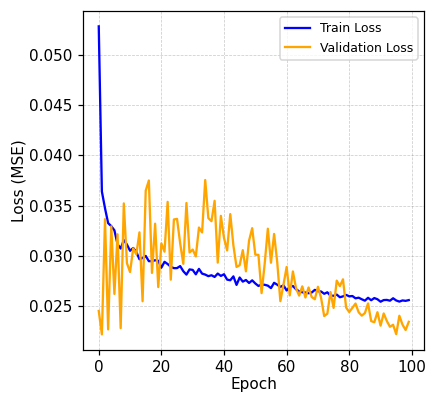
Using FRET assays, HLA alleles were grouped by isoLG adduct presentation: high (red), medium (orange), and low (blue). Notably, baseline FRET correlated with tBHP-stimulated IsoLG-adduct levels (Pearson r = 0.907), suggesting the presence of pre-existing IsoLG adducts (Figure 1A). After 100 training epochs (cycles), the model’s prediction error (loss) stabilized below 0.026 (Figure 1B). On the test set, the model’s predictions were typically within 13-16% of the normalized log(IC50) value (RMSE = 0.155; MAE = 0.134).
Conclusions
This study establishes a framework combining empirical binding assays, structural modeling, and graph-based deep learning for immunogenicity prediction in hypertension. The architecture shows promise in identifying relevant interaction sites between peptides and the HLA binding groove. Potent IsoLG-adduct presenting HLAs may represent a high-risk group for immune-mediated hypertension and related diseases.
Isolevuglandin (IsoLG)-adducted peptides contribute to hypertension by enhancing class I human leukocyte antigen (HLA)-mediated T cell activation. The capacity of different HLA alleles to present IsoLG-modified peptides varies, influencing immune activation and disease progression. To elucidate the molecular determinants of peptide-HLA (pHLA) interactions, we combined cell-based peptide binding assays, structural modeling, and deep learning to predict pHLA binding affinity (IC50) and the presentation of IsoLG-modified epitopes.
Methods
IsoLG presentation was quantified in K562 cells, each transfected to express a single HLA (45 total, covering 90% of the US population), using flow cytometry-based Förster resonance energy transfer (FRET), which measures peptide-HLA proximity via donor-acceptor fluorescence. In computational studies, we filtered experimental pHLA binding data from the Immune Epitope Databank (IEDB) and modeled 49,268 structures using Rosetta and AlphaFold. Graph embeddings were constructed for pHLA complexes and unbound HLA alleles, incorporating per-residue and pairwise binding energies, and IC50 values. HLA embeddings were enriched with IsoLG FRET data to train a graph neural network, which combines information from both local molecular structure and broader interaction patterns using graph convolution and self-attention. The approach accommodates missing data and variable graph sizes using custom loss (error tracking) functions to manage outliers and incomplete measurements.
Results
Using FRET assays, HLA alleles were grouped by isoLG adduct presentation: high (red), medium (orange), and low (blue). Notably, baseline FRET correlated with tBHP-stimulated IsoLG-adduct levels (Pearson r = 0.907), suggesting the presence of pre-existing IsoLG adducts (Figure 1A). After 100 training epochs (cycles), the model’s prediction error (loss) stabilized below 0.026 (Figure 1B). On the test set, the model’s predictions were typically within 13-16% of the normalized log(IC50) value (RMSE = 0.155; MAE = 0.134).
Conclusions
This study establishes a framework combining empirical binding assays, structural modeling, and graph-based deep learning for immunogenicity prediction in hypertension. The architecture shows promise in identifying relevant interaction sites between peptides and the HLA binding groove. Potent IsoLG-adduct presenting HLAs may represent a high-risk group for immune-mediated hypertension and related diseases.
More abstracts on this topic:
A Deep Learning Topic Analysis Approach for Enhancing Risk Assessment in Heart Failure Using Unstructured Clinical Notes
Adejumo Philip, Pedroso Aline, Khera Rohan
22q11 Deletion Syndrome: A Potenitial Risk Factor For Left Pulmonary Artery Hypoplasia and Need For Intervention in Patients With Congeital Heart DiseaseOliver Shannon, Ward Cameron