Final ID: MP2345
Multi-Omic Machine Learning for Predicting Outcomes in Heart Failure with Preserved Ejection Fraction (HFpEF): Insights from the UK Biobank
Abstract Body (Do not enter title and authors here): Background:
Heart failure with preserved ejection fraction (HFpEF) is a syndrome that may manifest differently in different people, and doctors lack clear tools for predicting how it will progress or how well medications will work. Traditional clinical risk assessments fail to capture the molecular complexity of HFpEF. Using machine learning (ML) to aggregate multi-omic data is a potential strategy for improving risk classification and discovering new biological subtypes.
Objective:
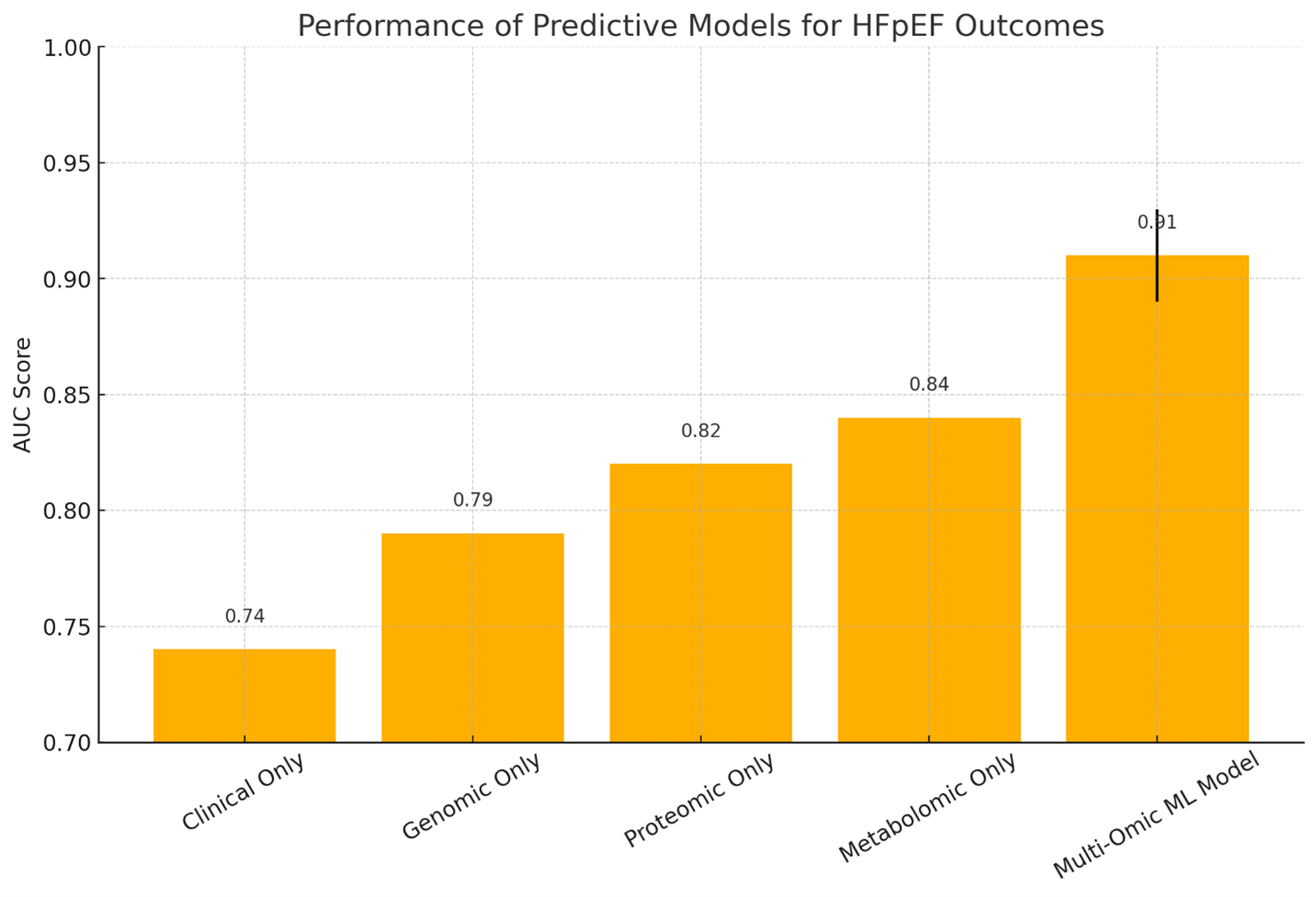
To develop and validate a multi-omic ML model that integrates clinical variables, genomic, proteomic, and metabolomic data to predict 1-year mortality and heart failure hospitalization in HFpEF patients.
Methods:
We analyzed data from 1,802 UK Biobank participants with validated HFpEF (LVEF > 50%). The patients' healthcare records, echocardiograms, genotyping arrays, proteomic profiles (Olink platform), and metabolomic profiles (Nightingale Health) were all connected. The main outcome was either death from any cause or hospitalization for heart failure within one year after the baseline evaluation. We built a machine learning pipeline with 10-fold cross-validation that used the XGBoost and random forest algorithms. We used recursive feature elimination and SHAP (Shapley Additive Explanations) to find features and make the model easier to understand.
Results:
The integrated multi-omic ML model had an AUC of 0.91 (95% CI: 0.88-0.93), significantly outperforming models based only on clinical factors (AUC 0.74) or individual omic layers (genomic: 0.79; proteomic: 0.82; metabolomic: 0.84). Key predictors were NT-proBNP, IL-6, GDF-15, branched-chain amino acids, and SNPs in myocardial fibrosis genes (TITIN, COL1A1). SHAP analysis revealed different high-risk molecular clusters, supporting the hypothesis of physiologically driven HFpEF subphenotypes.
Conclusion:
This study shows that a multi-omic ML method outperforms existing clinical models for predicting outcomes in HFpEF. The ability to combine molecular and clinical data may pave the way for physiologically informed, personalized HFpEF therapy. Prospective validation and implementation studies are required to translate these results into clinical practice.
Heart failure with preserved ejection fraction (HFpEF) is a syndrome that may manifest differently in different people, and doctors lack clear tools for predicting how it will progress or how well medications will work. Traditional clinical risk assessments fail to capture the molecular complexity of HFpEF. Using machine learning (ML) to aggregate multi-omic data is a potential strategy for improving risk classification and discovering new biological subtypes.
Objective:
To develop and validate a multi-omic ML model that integrates clinical variables, genomic, proteomic, and metabolomic data to predict 1-year mortality and heart failure hospitalization in HFpEF patients.
Methods:
We analyzed data from 1,802 UK Biobank participants with validated HFpEF (LVEF > 50%). The patients' healthcare records, echocardiograms, genotyping arrays, proteomic profiles (Olink platform), and metabolomic profiles (Nightingale Health) were all connected. The main outcome was either death from any cause or hospitalization for heart failure within one year after the baseline evaluation. We built a machine learning pipeline with 10-fold cross-validation that used the XGBoost and random forest algorithms. We used recursive feature elimination and SHAP (Shapley Additive Explanations) to find features and make the model easier to understand.
Results:
The integrated multi-omic ML model had an AUC of 0.91 (95% CI: 0.88-0.93), significantly outperforming models based only on clinical factors (AUC 0.74) or individual omic layers (genomic: 0.79; proteomic: 0.82; metabolomic: 0.84). Key predictors were NT-proBNP, IL-6, GDF-15, branched-chain amino acids, and SNPs in myocardial fibrosis genes (TITIN, COL1A1). SHAP analysis revealed different high-risk molecular clusters, supporting the hypothesis of physiologically driven HFpEF subphenotypes.
Conclusion:
This study shows that a multi-omic ML method outperforms existing clinical models for predicting outcomes in HFpEF. The ability to combine molecular and clinical data may pave the way for physiologically informed, personalized HFpEF therapy. Prospective validation and implementation studies are required to translate these results into clinical practice.
More abstracts on this topic:
A Cardiac Targeting Peptide Linked to miRNA106a Targets and Suppresses Genes Known to Cause Heart Failure: Reversing Heart Failure at the Source
Lu Ming, Deng Claire, Taskintuna Kaan, Ahern Gerard, Yurko Ray, Islam Kazi, Zahid Maliha, Gallicano Ian
β1-adrenergic autoantibodies (β1-AA) augment macropinocytosis in CD4+ T cells, leading to the expansion of CD4+CD28− T cell subsets in heart failure.Sun Fei, Yao Junyan, Li Bingjie, Zhang Suli, Liu Huirong