Final ID: MP2323
Explainable AutoML-Derived XGBoost Model for One-Year Mortality Prediction in HFpEF Patients
Abstract Body (Do not enter title and authors here): Background: Heart failure with preserved ejection fraction (HFpEF) accounts for approximately half of all heart failure hospitalizations; yet, tools for individualized 1-year mortality risk stratification remain limited. We conducted a retrospective study to develop and validate a machine learning model using genetic programming and automated model search to predict 1-year all-cause mortality in HFpEF patients admitted for decompensated HF.
Methods: Electronic medical records from Mayo Clinic (2010-2020) were queried to identify 7,840 adult patients hospitalized with HF exacerbation and a left ventricular ejection fraction>50% on echocardiograms performed between six months pre-admission and seven days post-discharge. Demographic, clinical, laboratory, medication, comorbidity, and imaging data available at the time of index hospitalization were extracted. The cohort was split into 80% training (n=6,272) and 20% held-out test (n=1,568) sets. After standard preprocessing, we performed an evolutionary AutoML search. The final pipeline applied a RobustScaler, two sequential feature-union blocks, and an XGBoost classifier. Model performance on the test set was assessed by area under the receiver operating characteristic curve (AUC), Brier score, accuracy, precision, recall, and F1 score, each with bootstrap-derived 95% confidence intervals (CIs). For interpretability, SHAP (SHapley Additive exPlanations) values were computed using the full scaled training set as background.
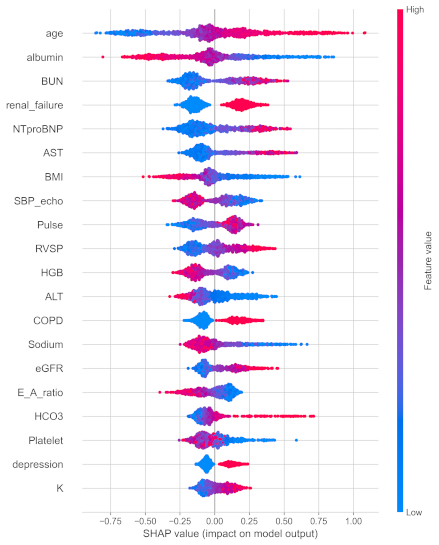
Results: On the held-out test set, the XGBoost-based model achieved an AUC of 0.7595 (95% CI: 0.7307-0.7870) and a Brier score of 0.1764 (95% CI: 0.1652-0.1890). Accuracy was 0.7406 (95% CI: 0.7143-0.7645), precision 0.6349 (95% CI: 0.5759-0.6942), recall 0.4135 (95% CI: 0.3636-0.4609), and F1 score 0.5008 (95% CI: 0.4529-0.5444). The global SHAP summary plot (Figure 1) ranked age, serum albumin, blood urea nitrogen (BUN), presence of renal failure, and NT-proBNP as the top 5 predictors.
Conclusions: In this large HFpEF cohort, an AutoML-derived XGBoost pipeline achieved robust discrimination (AUC≈0.76) and calibration (Brier≈0.18) for 1-year mortality prediction. SHAP-based explanations highlighted the importance of age, nutritional status (albumin), renal biomarkers (BUN, renal failure), and natriuretic peptides (NT-proBNP) as principal drivers of risk. These interpretable machine learning findings may guide personalized risk stratification and identify therapeutic targets in HFpEF.
Methods: Electronic medical records from Mayo Clinic (2010-2020) were queried to identify 7,840 adult patients hospitalized with HF exacerbation and a left ventricular ejection fraction>50% on echocardiograms performed between six months pre-admission and seven days post-discharge. Demographic, clinical, laboratory, medication, comorbidity, and imaging data available at the time of index hospitalization were extracted. The cohort was split into 80% training (n=6,272) and 20% held-out test (n=1,568) sets. After standard preprocessing, we performed an evolutionary AutoML search. The final pipeline applied a RobustScaler, two sequential feature-union blocks, and an XGBoost classifier. Model performance on the test set was assessed by area under the receiver operating characteristic curve (AUC), Brier score, accuracy, precision, recall, and F1 score, each with bootstrap-derived 95% confidence intervals (CIs). For interpretability, SHAP (SHapley Additive exPlanations) values were computed using the full scaled training set as background.
Results: On the held-out test set, the XGBoost-based model achieved an AUC of 0.7595 (95% CI: 0.7307-0.7870) and a Brier score of 0.1764 (95% CI: 0.1652-0.1890). Accuracy was 0.7406 (95% CI: 0.7143-0.7645), precision 0.6349 (95% CI: 0.5759-0.6942), recall 0.4135 (95% CI: 0.3636-0.4609), and F1 score 0.5008 (95% CI: 0.4529-0.5444). The global SHAP summary plot (Figure 1) ranked age, serum albumin, blood urea nitrogen (BUN), presence of renal failure, and NT-proBNP as the top 5 predictors.
Conclusions: In this large HFpEF cohort, an AutoML-derived XGBoost pipeline achieved robust discrimination (AUC≈0.76) and calibration (Brier≈0.18) for 1-year mortality prediction. SHAP-based explanations highlighted the importance of age, nutritional status (albumin), renal biomarkers (BUN, renal failure), and natriuretic peptides (NT-proBNP) as principal drivers of risk. These interpretable machine learning findings may guide personalized risk stratification and identify therapeutic targets in HFpEF.
More abstracts on this topic:
Age-Related Differences in Aortic Valve Calcium Progression and the Risk for Aortic Stenosis: Multi-Ethnic Study of Atherosclerosis
Marrero Natalie, Thanassoulis George, Rotter Jerome, Blaha Michael, Whelton Seamus, Jha Kunal, Grant Jelani, Razavi Alexander, Budoff Matthew, Shah Sanjiv, Blumenthal Roger, Post Wendy, Shaw Leslee
A Case of Concomitant Wild-Type Transthyretin and Systemic Light Chain Amyloidosis Involving Separate OrgansChiu Leonard, Afrough Aimaz, Nadeem Urooba, Jebakumar Deborah, Grodin Justin