Final ID: MP1415
Systematic Evaluation of Commercial and Open-source Large Language Models for Automated Adjudication of Clinical Indication from Cardiac Magnetic Resonance Imaging Reports
Abstract Body (Do not enter title and authors here): Background: Indications for cardiac magnetic resonance imaging (CMR) are often stored in heterogenous, unstructured reports. Manual adjudication of indications is time-consuming and requires domain expertise. Recent large language models (LLMs) have shown promise in complex clinical interpretation and categorization tasks. No prior study has systematically evaluated the ability of state-of-the-art (SOTA) LLMs to extract indications from raw CMR reports.
Research question: How well do SOTA open-source and commercial LLMs adjudicate clinical indications from real-world CMR reports?
Methods: We analyzed 486 CMR reports from a large academic center. Reports were de-identified using the Stanford-Penn-MIDRC deidentification tool, and ground-truth indications were annotated by a physician expert. 18 LLMs varying in accessibility (8 open-source, 10 commercial), parameter size (4 to 70 billion), and training corpus (general vs medical) were evaluated. For each report, LLMs were instructed to extract the top two possible indications (correct if either matched the ground-truth indication)—reflecting the fact that real-world indications can fall into more than 1 category—from ten possible categories: oncologic therapy toxicity, cardiomyopathy/elevated troponin, chest pain/dyspnea, arrythmia/abnormal ECG, cardiac mass/metastasis, thrombus, structural evaluation, pericarditis, risk stratification, or viability evaluation (ischemic).
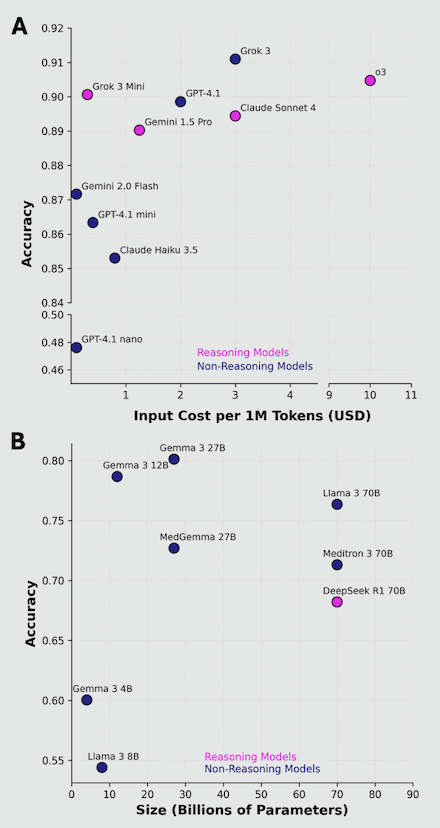
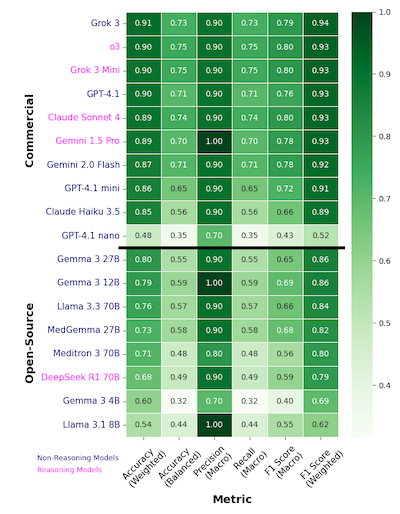
Results: Higher-cost commercial models (Spearman’s rank r = 0.683, p = 0.03) and larger-parameter open-source models (r = 0.307) exhibited better adjudication ability, Fig 1A, 1B. The best performing commercial LLMs performed markedly better than the top open-source LLMs (90% vs ~78% accuracy [acc]), Fig 2. Grok 3 (91% acc, 0.94 F1-score) and OpenAI o3 (90% acc, 0.93 F1) were the best models overall, and Gemma 3 27B was the best open-source LLM (80% acc, 0.86 F1), Fig 2. Reasoning models performed comparably to non-reasoning models, with Grok 3 mini having the best relative cost-vs-performance, Fig 1A, 2. Interestingly, medical LLMs performed worse than their generally pretrained counterparts (e.g., MedGemma 27B vs Gemma 3 27B), suggesting domain-specific pretraining may negatively affect adjudication ability, Fig 2.
Conclusion: Open-source and commercial LLMs demonstrate promise in automated, accurate extraction of indications from CMR reports. Our findings help clinician-researchers decide between LLMs for use-cases involving CMR reports.
Research question: How well do SOTA open-source and commercial LLMs adjudicate clinical indications from real-world CMR reports?
Methods: We analyzed 486 CMR reports from a large academic center. Reports were de-identified using the Stanford-Penn-MIDRC deidentification tool, and ground-truth indications were annotated by a physician expert. 18 LLMs varying in accessibility (8 open-source, 10 commercial), parameter size (4 to 70 billion), and training corpus (general vs medical) were evaluated. For each report, LLMs were instructed to extract the top two possible indications (correct if either matched the ground-truth indication)—reflecting the fact that real-world indications can fall into more than 1 category—from ten possible categories: oncologic therapy toxicity, cardiomyopathy/elevated troponin, chest pain/dyspnea, arrythmia/abnormal ECG, cardiac mass/metastasis, thrombus, structural evaluation, pericarditis, risk stratification, or viability evaluation (ischemic).
Results: Higher-cost commercial models (Spearman’s rank r = 0.683, p = 0.03) and larger-parameter open-source models (r = 0.307) exhibited better adjudication ability, Fig 1A, 1B. The best performing commercial LLMs performed markedly better than the top open-source LLMs (90% vs ~78% accuracy [acc]), Fig 2. Grok 3 (91% acc, 0.94 F1-score) and OpenAI o3 (90% acc, 0.93 F1) were the best models overall, and Gemma 3 27B was the best open-source LLM (80% acc, 0.86 F1), Fig 2. Reasoning models performed comparably to non-reasoning models, with Grok 3 mini having the best relative cost-vs-performance, Fig 1A, 2. Interestingly, medical LLMs performed worse than their generally pretrained counterparts (e.g., MedGemma 27B vs Gemma 3 27B), suggesting domain-specific pretraining may negatively affect adjudication ability, Fig 2.
Conclusion: Open-source and commercial LLMs demonstrate promise in automated, accurate extraction of indications from CMR reports. Our findings help clinician-researchers decide between LLMs for use-cases involving CMR reports.
More abstracts on this topic:
4D Flow MRI Allows for Enhanced Characterization of Aortic Regurgitation
Avgousti Harris, Johnson Ethan, Berhane Haben, Thomas James, Allen Bradley, Markl Michael, Appadurai Vinesh
A REPORT OF >2000 CONSECUTIVE NON-SELECTED PATIENTS UNDERGOING IMAGING WITH PACEMAKER'S AND ICD'S IN THE MRI ENVIRONMENT; THE ADDITIVE VALUE OF A LARGE, PROSPECTIVE >20-YEAR OBSERVATIONAL STUDYBiederman Robert, Shah Moneal, Lombardi Rich, Boltralik Mark, Mangini Francesco, Doyle Mark