Final ID: Su2015
A novel deep learning framework identified associated genes and Interpretable deep learning translation of GWAS findings for drug repurposing in Atrial Fibrillation
Abstract Body (Do not enter title and authors here): Introduction: Translating human genetic findings, such as genome-wide association studies (GWAS) to pathobiology and the discovery of therapeutic target remains a challenge for Atrial Fibrillation (AF). We developed a novel network topology-based deep learning framework to identify disease-associated genes (TAGX). TAGX is a computational framework that encodes protein-protein interactomes (PPIs) enriched with cliques (i.e., a set of nodes such that every pair of nodes in the set is connected by an edge) via three views (i.e., local, global and hypergraph). In the downstream analysis, the learned node/protein embeddings are combined with AF associated genes with genetic evidence to prioritize additional risk genes.
Aims: To identify potential AF related genes (afRG) and repurposable drug candidates using our deep learning framework.
Methods: We previously built a comprehensive human PPI containing 351,444 unique interactions and 17,706 proteins. The existence of cliques in PPIs increase the representational challenge, therefore TAGX learns each node’s representation in the entire PPI by considering three structured views: local (i.e., up to 2nd order neighbors of each node), global (i.e., modelling the interactions between each node and all other nodes in the PPI network), and hypergraph (i.e., how frequently two nodes are in the same clique). By combining the protein embedding learned by TAGX with colocalized signals between AF GWAS loci and heart quantitative trait loci (QTLs), we prioritized afRG. Then, using network proximity approaches to evaluate the closest distance between afRGs and a drug’s targets within the human PPI, we computationally predicted drugs for AF.
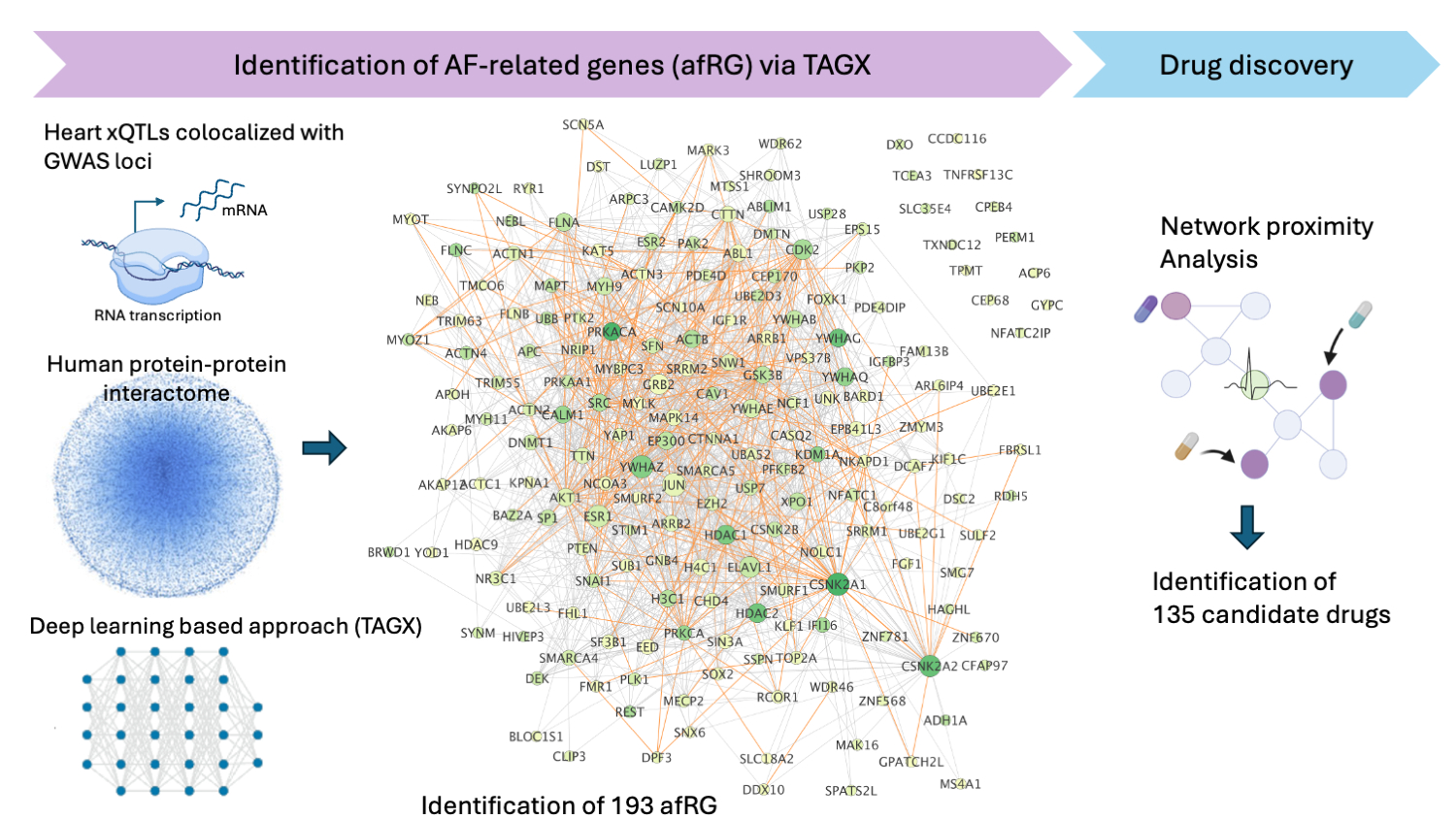
Results: We first identified 208 expression QTLs, 19 splicing QTLs, and 2 protein QTLs that are colocalized with AF GWAS hits. Via TAGX, we identified 193 afRGs (Figure), including known AF related genes such as MYOZ1, CAV1, SYNPO2L, TTN or SCN5A. Among the 193 predicted afRGs, >50% of the genes were reported as AF associated genes previously. Using the closest-based network proximity approach, we computationally identified 135 candidate drugs spanning 14 first-level anatomical therapeutic chemical classes. These included drugs reportedly potentially treating AF, such as Telmisartan (Z=-2.52), Pioglitazone (Z=-2.77), or Fenofibrate (Z=-2.69).
Conclusion: Using a novel deep learning methodology that utilized GWAS and QTL findings, we identified risk genes and repurposing drug candidates for AF.
Aims: To identify potential AF related genes (afRG) and repurposable drug candidates using our deep learning framework.
Methods: We previously built a comprehensive human PPI containing 351,444 unique interactions and 17,706 proteins. The existence of cliques in PPIs increase the representational challenge, therefore TAGX learns each node’s representation in the entire PPI by considering three structured views: local (i.e., up to 2nd order neighbors of each node), global (i.e., modelling the interactions between each node and all other nodes in the PPI network), and hypergraph (i.e., how frequently two nodes are in the same clique). By combining the protein embedding learned by TAGX with colocalized signals between AF GWAS loci and heart quantitative trait loci (QTLs), we prioritized afRG. Then, using network proximity approaches to evaluate the closest distance between afRGs and a drug’s targets within the human PPI, we computationally predicted drugs for AF.
Results: We first identified 208 expression QTLs, 19 splicing QTLs, and 2 protein QTLs that are colocalized with AF GWAS hits. Via TAGX, we identified 193 afRGs (Figure), including known AF related genes such as MYOZ1, CAV1, SYNPO2L, TTN or SCN5A. Among the 193 predicted afRGs, >50% of the genes were reported as AF associated genes previously. Using the closest-based network proximity approach, we computationally identified 135 candidate drugs spanning 14 first-level anatomical therapeutic chemical classes. These included drugs reportedly potentially treating AF, such as Telmisartan (Z=-2.52), Pioglitazone (Z=-2.77), or Fenofibrate (Z=-2.69).
Conclusion: Using a novel deep learning methodology that utilized GWAS and QTL findings, we identified risk genes and repurposing drug candidates for AF.
More abstracts on this topic:
A Deep Learning Topic Analysis Approach for Enhancing Risk Assessment in Heart Failure Using Unstructured Clinical Notes
Adejumo Philip, Pedroso Aline, Khera Rohan
A Novel Variant in GNB2 as a Cause of Sick Sinus SyndromeBulut Aybike, Karacan Mehmet, Saygili E. Alper, Pirli Dogukan, Aydin Eylul, Ozdemir Ozkan, Balci Nermin, Alanay Yasemin, Bilguvar Kaya, Akgun Dogan Ozlem