Final ID: MP2346
Phenotyping Cardiac Surgery Patients Using Retrieval-Augmented Large Language Models
Abstract Body (Do not enter title and authors here): Introduction:
Large Language Models (LLMs) are powerful tools for text extraction, but their tendency to hallucinate limits their reliability in clinical domains. We present a novel application of retrieval-augmented generation (RAG) to reduce hallucinations. Our approach restricts context to short, high-similarity segments within cardiac imaging reports, enabling more focused, conservative inference. We applied RAG to extract echocardiographic features from intraoperative transesophageal echocardiography (TEE) reports in a mixed cardiac surgery population to identify distinct patient phenotypes.
Hypothesis:
We hypothesized that RAG would outperform direct LLM querying in extracting key echocardiographic features by reducing hallucinations. We aimed to group patients into clinically meaningful clusters by their echocardiographic features.
Methods:
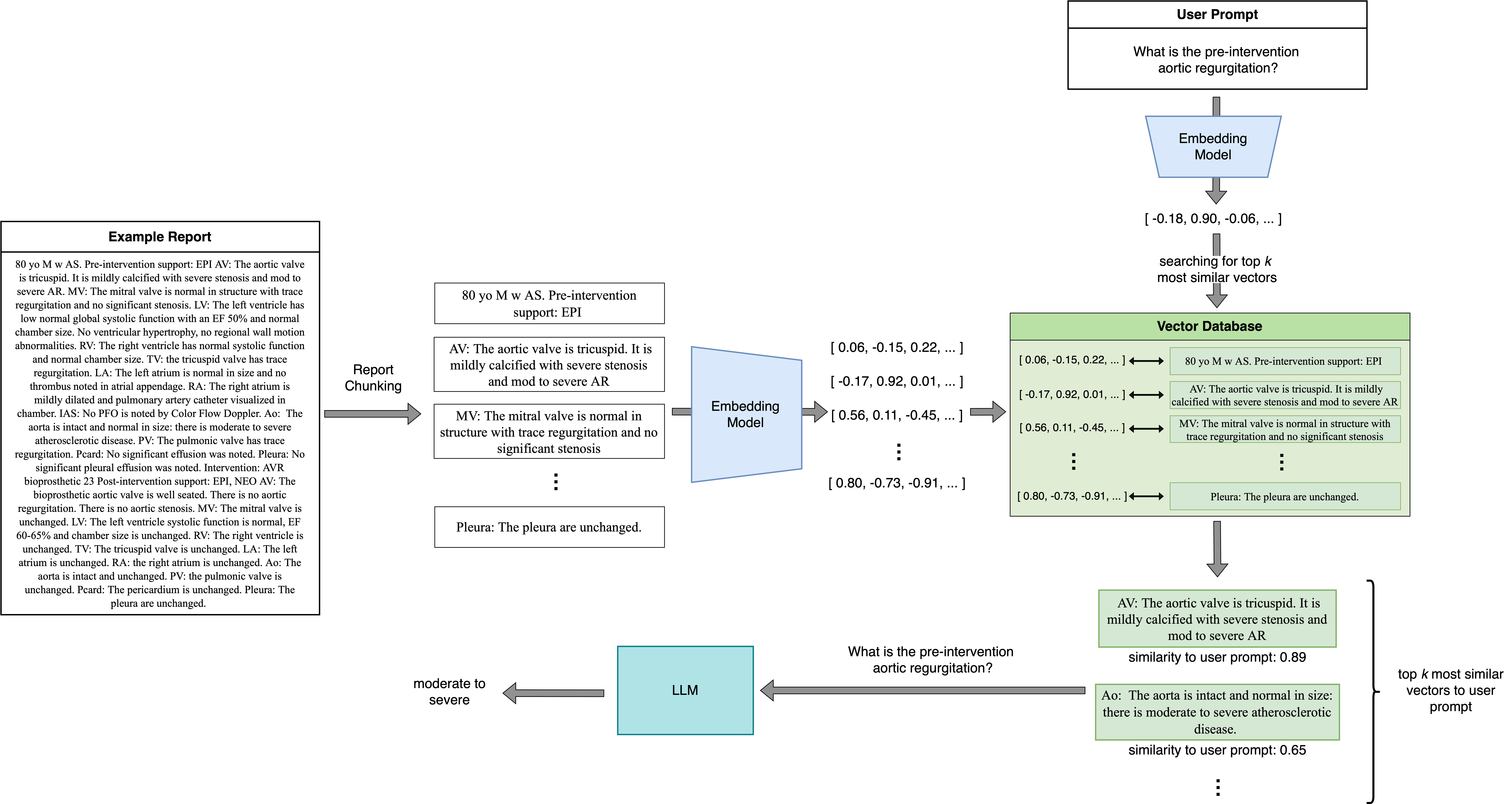
We developed a RAG pipeline that restricts LLM input to the most semantically relevant portions of TEE reports (Figure 1). We validated this pipeline on 500 manually labeled reports, extracting pre- and post-intervention left ventricular ejection fraction (LVEF), tricuspid regurgitation (TR), and right ventricular systolic function (RVSF), as well as pre-intervention aortic stenosis (AS), aortic regurgitation (AR), and mitral regurgitation (MR). RAG performance was compared to direct querying on these validation reports. Next, the pipeline was scaled to 7106 TEE reports to extract the features and intervention types. Patients were clustered using k-means, and each cluster’s characteristics were analyzed.
Results:
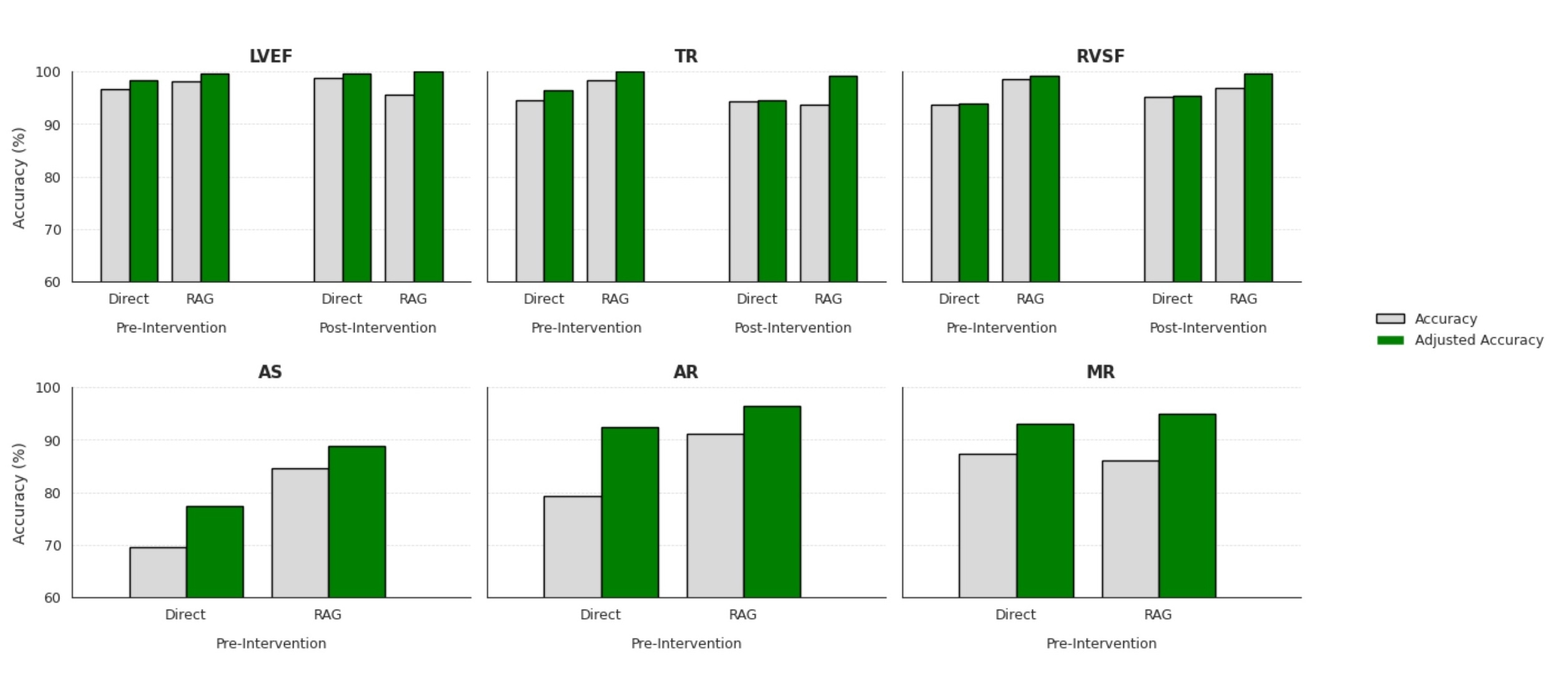
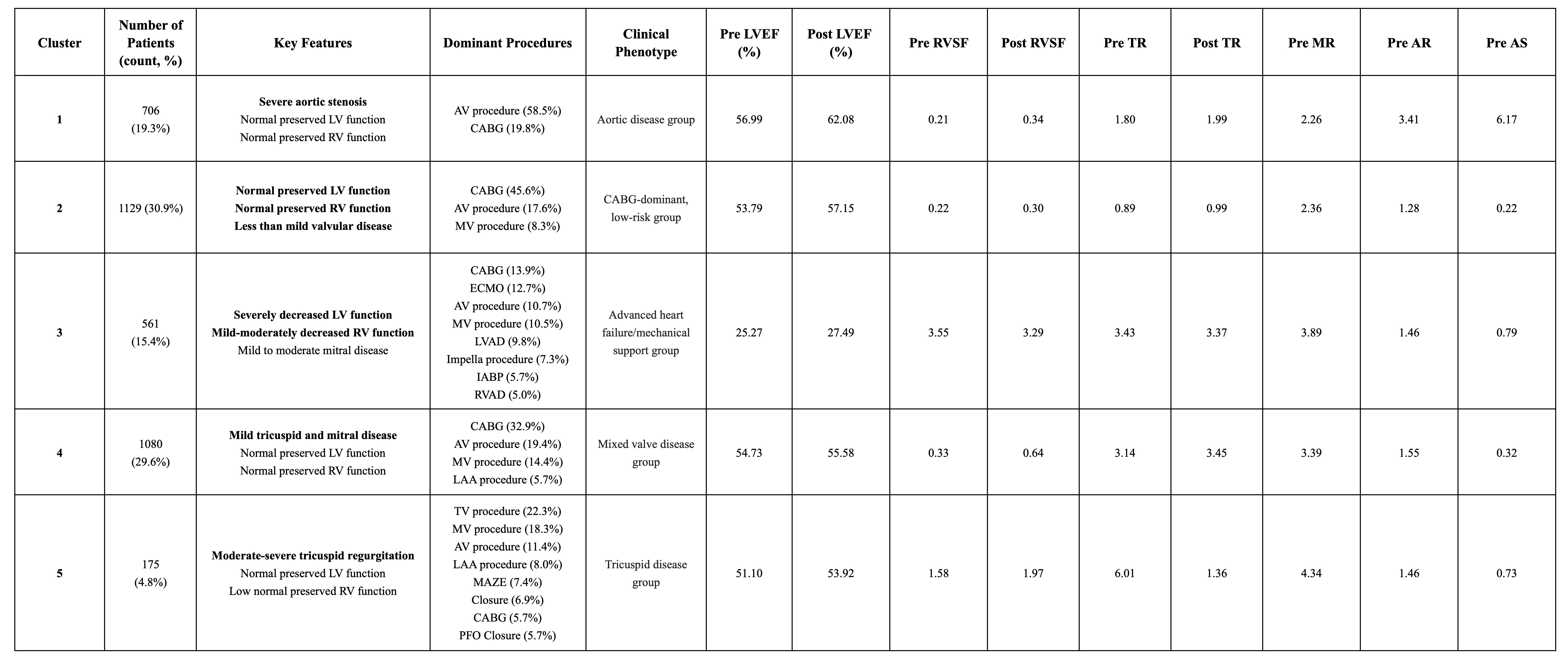
RAG’s conservative behavior—favoring “not found” over potential fabrications—resulted in fewer hallucinations compared to direct LLM queries (Figure 2): RAG improved adjusted accuracy across all validation features (LVEF pre: +1.24%, LVEF post: +0.47%, TR pre: +3.64%, TR post: +4.67%, RVSF pre: +5.31%, RVSF post: +4.33%, AS pre: +11.44%, AR pre: +3.93%, MR pre: +1.94%). Clustering revealed five distinct phenotypes: (1) an aortic disease group, (2) a CABG-dominant low risk group, (3) an advanced heart failure group, (4) a mixed valve disease group, and (5) a tricuspid disease group (Table 1).
Conclusions:
Our RAG pipeline improves the reliability of LLM-based clinical data extraction from TEE reports, enabling large-scale phenotyping of heterogeneous cardiac surgery populations. This approach has potential applications for personalized risk stratification and targeted clinical decision support in cardiac surgery.
Large Language Models (LLMs) are powerful tools for text extraction, but their tendency to hallucinate limits their reliability in clinical domains. We present a novel application of retrieval-augmented generation (RAG) to reduce hallucinations. Our approach restricts context to short, high-similarity segments within cardiac imaging reports, enabling more focused, conservative inference. We applied RAG to extract echocardiographic features from intraoperative transesophageal echocardiography (TEE) reports in a mixed cardiac surgery population to identify distinct patient phenotypes.
Hypothesis:
We hypothesized that RAG would outperform direct LLM querying in extracting key echocardiographic features by reducing hallucinations. We aimed to group patients into clinically meaningful clusters by their echocardiographic features.
Methods:
We developed a RAG pipeline that restricts LLM input to the most semantically relevant portions of TEE reports (Figure 1). We validated this pipeline on 500 manually labeled reports, extracting pre- and post-intervention left ventricular ejection fraction (LVEF), tricuspid regurgitation (TR), and right ventricular systolic function (RVSF), as well as pre-intervention aortic stenosis (AS), aortic regurgitation (AR), and mitral regurgitation (MR). RAG performance was compared to direct querying on these validation reports. Next, the pipeline was scaled to 7106 TEE reports to extract the features and intervention types. Patients were clustered using k-means, and each cluster’s characteristics were analyzed.
Results:
RAG’s conservative behavior—favoring “not found” over potential fabrications—resulted in fewer hallucinations compared to direct LLM queries (Figure 2): RAG improved adjusted accuracy across all validation features (LVEF pre: +1.24%, LVEF post: +0.47%, TR pre: +3.64%, TR post: +4.67%, RVSF pre: +5.31%, RVSF post: +4.33%, AS pre: +11.44%, AR pre: +3.93%, MR pre: +1.94%). Clustering revealed five distinct phenotypes: (1) an aortic disease group, (2) a CABG-dominant low risk group, (3) an advanced heart failure group, (4) a mixed valve disease group, and (5) a tricuspid disease group (Table 1).
Conclusions:
Our RAG pipeline improves the reliability of LLM-based clinical data extraction from TEE reports, enabling large-scale phenotyping of heterogeneous cardiac surgery populations. This approach has potential applications for personalized risk stratification and targeted clinical decision support in cardiac surgery.
More abstracts on this topic:
Angiography-derived FFR-Guided Coronary Artery Bypass Grafting for Patients undergoing Valve Surgery with Concomitant Coronary Artery Disease (FAVOR 4-QVAS): a Randomized Trial
A Case of Possible IgG4-Related Constrictive Pericarditis Masquerading as Idiopathic Pericarditis: A Rare and Elusive Diagnosis
Zhu Yunpeng, Guo Zhigang, Zhu Dan, Zhang Xiquan, Chen Liangwan, Redfors Bjorn, Sandner Sigrid, Gaudino Mario, Tu Shengxian, Zhao Qiang, Cheng Zhaoyun, Zhao Yuan, Zhang Wei, Han Lin, Zhang Chengxin, Yang Sumin, Ma Liang, Qiao Chenhui
A Case of Possible IgG4-Related Constrictive Pericarditis Masquerading as Idiopathic Pericarditis: A Rare and Elusive Diagnosis
Nandyal Shreyas, Sharma Bharosa, Gajjar Rohan, Varma Revati, Ezegwu Olisa, Amdetison Gedion Yilma, Tottleben Jon