Final ID: MP371
A novel XAI framework for explainable AI-ECG using generative counterfactual-based XAI
Abstract Body (Do not enter title and authors here): Background: To ensure clinical adoption of artificial intelligence (AI)-electrocardiogram (ECG), it is essential to understand why the AI makes certain predictions. Traditional explainable AI (XAI) methods like saliency maps highlight important ECG segments but often fall short in explaining how specific morphology or rhythm changes influence predictions.
Purpose: We propose a generative counterfactual-based explainability (GCX) framework that simulates “how changes” in ECG waveforms affect AI-ECG predictions.
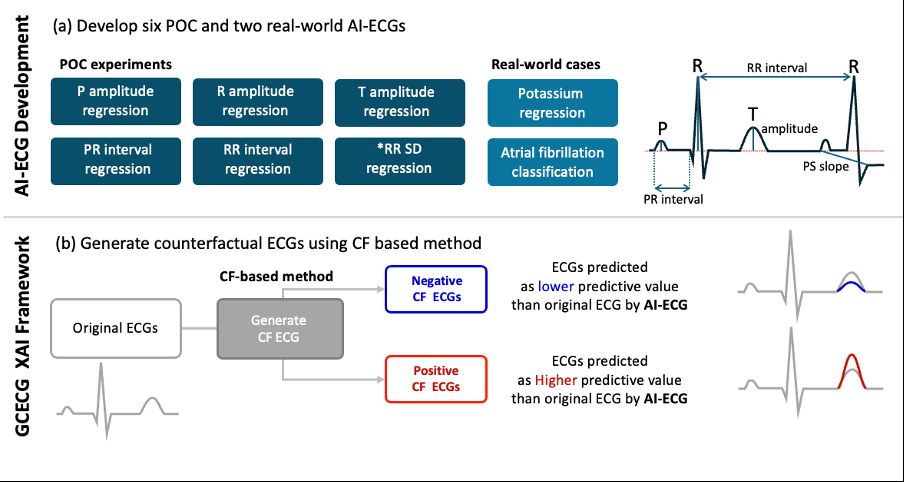
Methods: We developed eight AI-ECGs: six for ECG feature regression (P wave, R wave, and T wave amplitudes; PR interval; RR interval; RR variability), one for serum potassium regression, and one for atrial fibrillation (AF) classification. Models were trained using PTB-XL or MIMIC-IV datasets. We applied the GCX framework using a generative AI model (StyleGAN2) to create "positive counterfactual ECGs" (with higher predicted values) and "negative counterfactual ECGs" (with lower predicted values). (Figure 1) By comparing these to the original ECGs, we identified waveform changes that influenced AI predictions. To evaluate consistency, we performed paired t-tests between original and counterfactual ECG features.
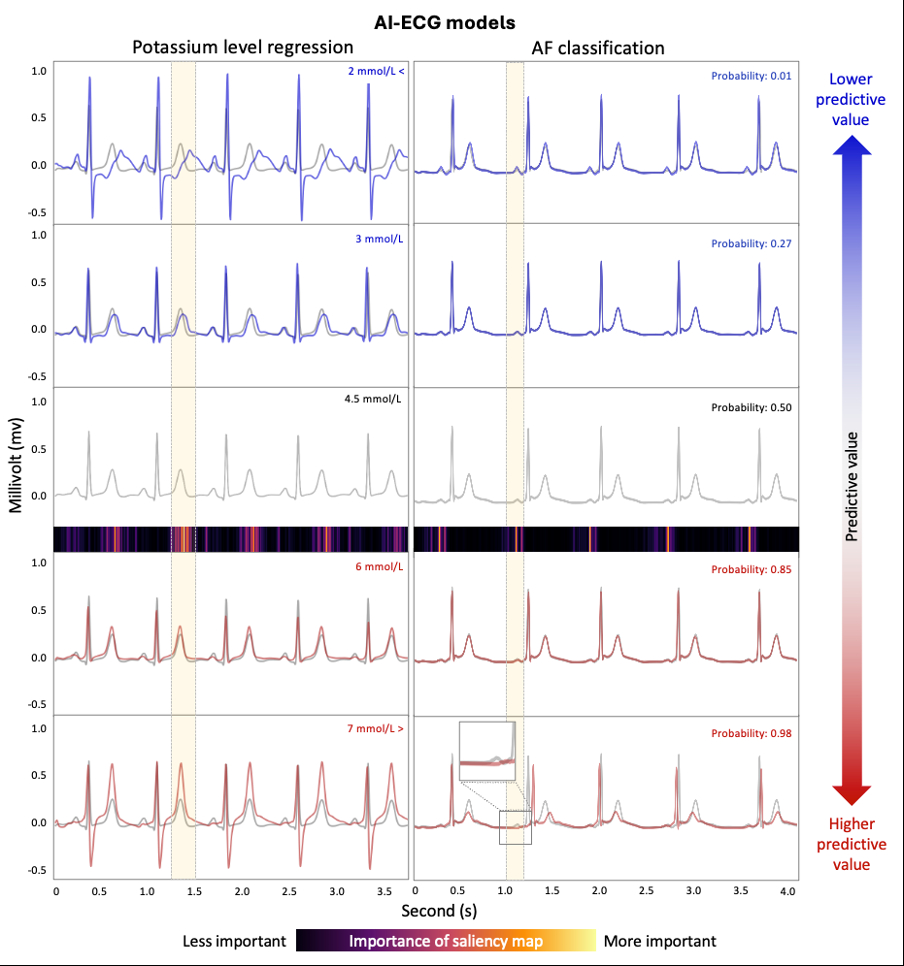
Results: In all six feature regression models, positive counterfactual ECGs consistently showed increased amplitudes (P, R, T waves), prolonged RR intervals, and greater RR variability. (Figure 2) Negative counterfactuals showed the opposite. For the potassium regression model, positive counterfactual ECGs simulating hyperkalemia displayed increased T wave amplitude, PR prolongation, and QRS widening. Negative counterfactuals resembling hypokalemia showed decreased T wave amplitude, increased P wave amplitude, ST depression, and prominent U waves. In the AF classification model, positive counterfactuals revealed progressive loss of P waves and increased rhythm irregularity. (Figure 2, 3) Across all models, feature changes between original and counterfactual ECGs were statistically significant (paired t-test, p < 0.01), confirming the robustness of the explanations.
Conclusion: The GCX framework revealed how AI-ECG models determine predictions based on specific waveform and rhythm changes. These explanations aligned well with established electrophysiological knowledge. By clarifying the "reasoning process", GCX offers a clinically meaningful path toward safer, more transparent, and interpretable deployment of AI-ECG in real-world practice.
Purpose: We propose a generative counterfactual-based explainability (GCX) framework that simulates “how changes” in ECG waveforms affect AI-ECG predictions.
Methods: We developed eight AI-ECGs: six for ECG feature regression (P wave, R wave, and T wave amplitudes; PR interval; RR interval; RR variability), one for serum potassium regression, and one for atrial fibrillation (AF) classification. Models were trained using PTB-XL or MIMIC-IV datasets. We applied the GCX framework using a generative AI model (StyleGAN2) to create "positive counterfactual ECGs" (with higher predicted values) and "negative counterfactual ECGs" (with lower predicted values). (Figure 1) By comparing these to the original ECGs, we identified waveform changes that influenced AI predictions. To evaluate consistency, we performed paired t-tests between original and counterfactual ECG features.
Results: In all six feature regression models, positive counterfactual ECGs consistently showed increased amplitudes (P, R, T waves), prolonged RR intervals, and greater RR variability. (Figure 2) Negative counterfactuals showed the opposite. For the potassium regression model, positive counterfactual ECGs simulating hyperkalemia displayed increased T wave amplitude, PR prolongation, and QRS widening. Negative counterfactuals resembling hypokalemia showed decreased T wave amplitude, increased P wave amplitude, ST depression, and prominent U waves. In the AF classification model, positive counterfactuals revealed progressive loss of P waves and increased rhythm irregularity. (Figure 2, 3) Across all models, feature changes between original and counterfactual ECGs were statistically significant (paired t-test, p < 0.01), confirming the robustness of the explanations.
Conclusion: The GCX framework revealed how AI-ECG models determine predictions based on specific waveform and rhythm changes. These explanations aligned well with established electrophysiological knowledge. By clarifying the "reasoning process", GCX offers a clinically meaningful path toward safer, more transparent, and interpretable deployment of AI-ECG in real-world practice.
More abstracts on this topic:
12-lead electrocardiograms predict adverse cardiovascular outcomes of emergency department patients
Haimovich Julian, Kolossvary Marton, Alam Ridwan, Padros I Valls Raimon, Lu Michael, Aguirre Aaron
9-Year Longitudinal Assessment of the 12-lead Electrocardiogram of Volunteer FirefightersBae Alexander, Dzikowicz Dillon, Lai Chi-ju, Brunner Wendy, Krupa Nicole, Carey Mary, Tam Wai Cheong, Yu Yichen