Final ID: Mo4049
Proteome-wide analysis of cardiac structure, function, and heart failure phenotypes using Mendelian Randomization
Abstract Body (Do not enter title and authors here): Introduction: Heart failure (HF) is a complex, heterogeneous disease. Identifying circulating proteins linked to cardiac structure, function, and HF subtypes may reveal new therapeutic targets. Mendelian randomization (MR) using protein quantitative trait loci (pQTL) and cardiac magnetic resonance imaging (cMRI) phenotypes enables systematic evaluation of proteomic influences on cardiac traits and HF subtypes.
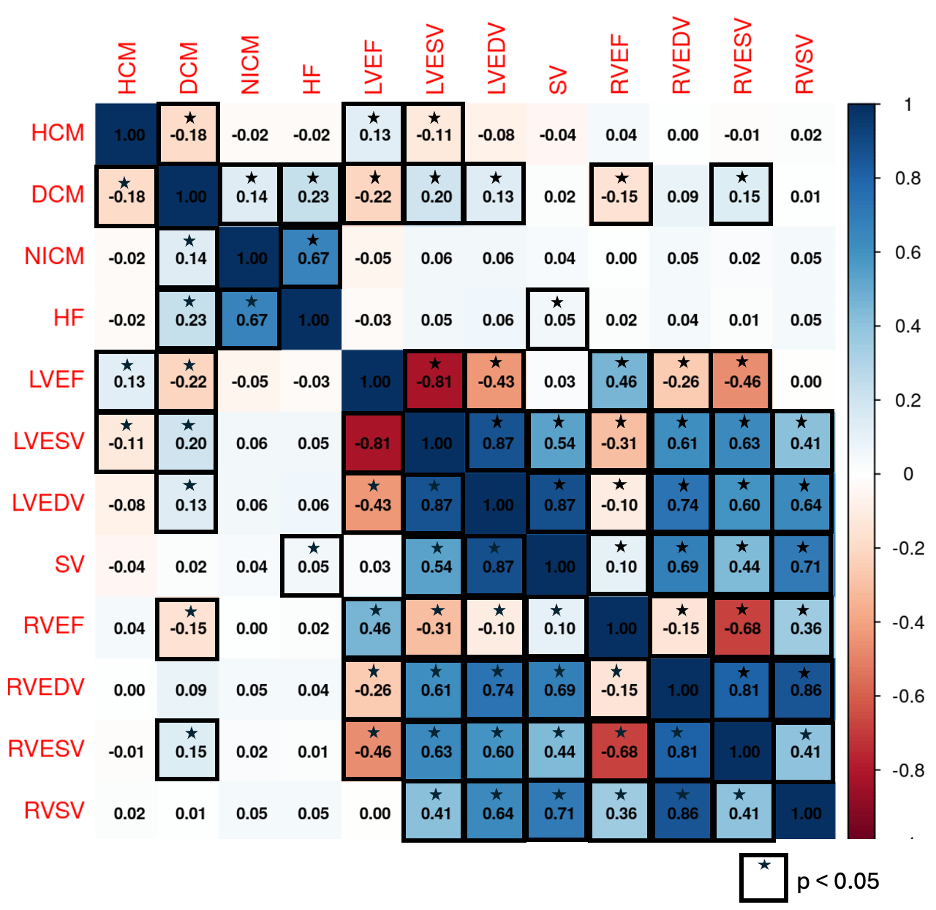
Methods: We performed proteome-wide univariable MR analyses to investigate potential causal effects of 1,715 serum proteins on eight cMRI traits of left and right ventricular function (LVEF, LVESV, LVEDV, SV, RVEF, RVESV, RVEDV, and RVSV) and four HF subtypes (all-cause HF, non-ischemic cardiomyopathy [NICM], dilated cardiomyopathy [DCM], and hypertrophic cardiomyopathy [HCM]). Genetic instruments were derived from pQTL data in 34,557 UK Biobank participants. Outcome data included cMRI traits (36,041 individuals), HF/NICM (153,174 cases), HCM (5,900 cases) and DCM (9,365 cases). MR was performed using the inverse-variance weighted method. We visualized significant (q < 0.05) associations across traits to highlight differences and overlap, and used a heatmap to depict Pearson correlations of z-score MR estimates across traits.
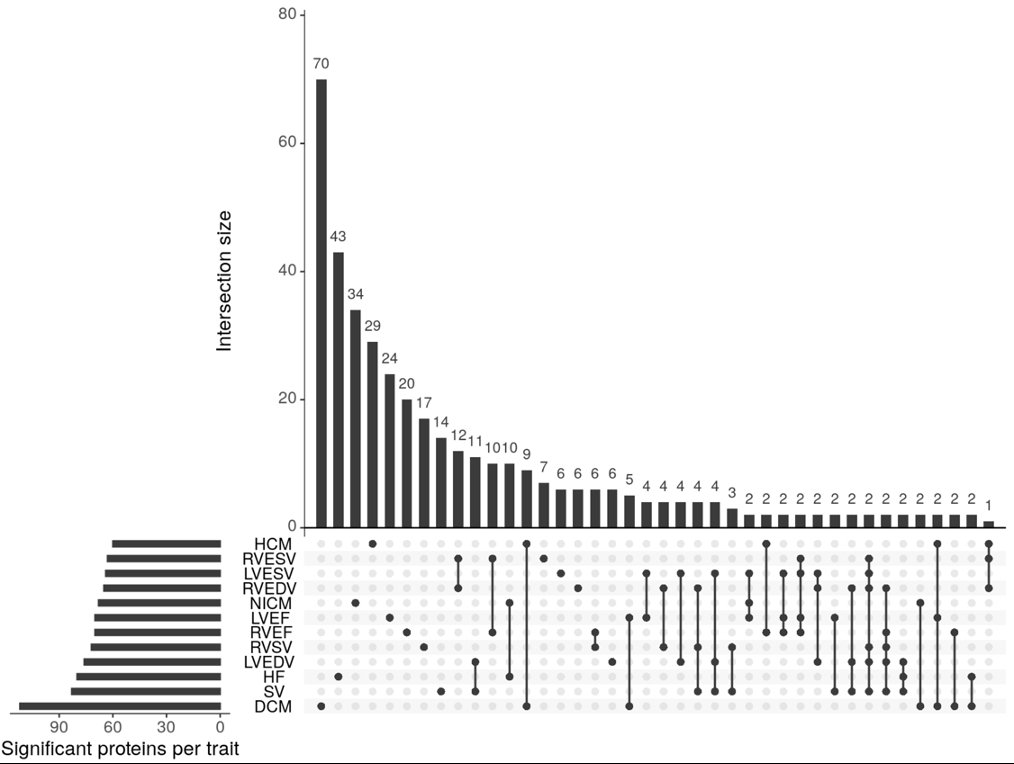
Results: Proteome-wide MR identified 60-112 significant associations per trait. Visualization revealed both distinct and overlapping protein signatures, suggesting pleiotropic influences on cardiac remodeling and HF phenotypes. Trait-trait correlation analysis revealed significant positive correlations between protein effects on NICM and HF (r=0.67, p < 0.05), while protein effects on HCM and DCM displayed an inverse relationship (r=–0.18, p < 0.05), indicating divergent proteomic pathways in these cardiomyopathies. Ventricular volume traits (LVESV, LVEDV, RVEDV, RVESV) were highly intercorrelated (r>0.6), indicating proteomic drivers of ventricular remodeling may be conserved. Pathway analysis of proteins significantly associated with ≥4 traits revealed enrichment in protein processing, immune response, and lipid transport. Proteins associated with ≥6 traits included known and novel markers: SPON1, ABO, CNTN2, ENG, PDCD6, PLA2G2A, TGFBI, TLR1, TPSD1, and VSNL1.
Conclusion: This analysis highlights both distinct and shared circulating proteins associated with cardiac structure, function, and HF phenotypes. Further exploration of these proteins and pathways could inform novel therapeutic strategies conserved across HF etiologies.
Methods: We performed proteome-wide univariable MR analyses to investigate potential causal effects of 1,715 serum proteins on eight cMRI traits of left and right ventricular function (LVEF, LVESV, LVEDV, SV, RVEF, RVESV, RVEDV, and RVSV) and four HF subtypes (all-cause HF, non-ischemic cardiomyopathy [NICM], dilated cardiomyopathy [DCM], and hypertrophic cardiomyopathy [HCM]). Genetic instruments were derived from pQTL data in 34,557 UK Biobank participants. Outcome data included cMRI traits (36,041 individuals), HF/NICM (153,174 cases), HCM (5,900 cases) and DCM (9,365 cases). MR was performed using the inverse-variance weighted method. We visualized significant (q < 0.05) associations across traits to highlight differences and overlap, and used a heatmap to depict Pearson correlations of z-score MR estimates across traits.
Results: Proteome-wide MR identified 60-112 significant associations per trait. Visualization revealed both distinct and overlapping protein signatures, suggesting pleiotropic influences on cardiac remodeling and HF phenotypes. Trait-trait correlation analysis revealed significant positive correlations between protein effects on NICM and HF (r=0.67, p < 0.05), while protein effects on HCM and DCM displayed an inverse relationship (r=–0.18, p < 0.05), indicating divergent proteomic pathways in these cardiomyopathies. Ventricular volume traits (LVESV, LVEDV, RVEDV, RVESV) were highly intercorrelated (r>0.6), indicating proteomic drivers of ventricular remodeling may be conserved. Pathway analysis of proteins significantly associated with ≥4 traits revealed enrichment in protein processing, immune response, and lipid transport. Proteins associated with ≥6 traits included known and novel markers: SPON1, ABO, CNTN2, ENG, PDCD6, PLA2G2A, TGFBI, TLR1, TPSD1, and VSNL1.
Conclusion: This analysis highlights both distinct and shared circulating proteins associated with cardiac structure, function, and HF phenotypes. Further exploration of these proteins and pathways could inform novel therapeutic strategies conserved across HF etiologies.
More abstracts on this topic:
3D Chromatin Architectures and Transcription Regulation in Diabetic Endothelial Dysfunction
Feng Yuliang, Cai Liuyang, Wang Yigang, Huang Wei, Jiang Lei
ADP-Ribosylation In a Mouse Model of Atherosclerosis: a Potential Novel Link Between Dyslipidemia and Inflammation in Cardiovascular DiseaseDelwarde Constance, Mlynarchik Andrew, Perez Katelyn, Campedelli Alesandra, Sonawane Abhijeet, Aikawa Elena, Singh Sasha, Aikawa Masanori, Santinelli Pestana Diego, Kasai Taku, Kuraoka Shiori, Nakamura Yuto, Okada Takeshi, Decano Julius, Chelvanambi Sarvesh, Ge Rile