Final ID: Su1035
Genetic evaluation of metabolic signatures associated with lipid-lowering medications
Abstract Body (Do not enter title and authors here): Introduction: Human genetic studies have identified hundreds of risk loci for circulating lipoproteins, motivating the development of new lipid-lowering medications. Although these targets influence lipids and metabolites through unique mechanisms, the extent to which they exert shared pleiotropic effects on other effectors of atherosclerotic cardiovascular disease remains uncertain. This study explored the lipid subfraction, cytokine, and metabolic signatures associated with lipid-lowering targets using Mendelian Randomization (MR).
Methods: The study focused on protein targets of novel lipid-lowering medications: PCSK9, APOB, LPA, LPL, and ANGPTL3. Instruments for PCSK9, LPL, and ANGPTL3 were derived from the UK Biobank Pharma Proteomics Project (54,219 participants), and for LPA and APOB from the Pan-UK Biobank (335,796 and 418,505 participants). These exposures were tested against outcomes including 40 cytokines (meta-analysis of 74,783 individuals), 249 lipid-subfraction measures (118,461 UK Biobank participants), and 1,400 metabolite measures (8,299 participants in the Canadian Longitudinal Study on Aging). Univariable MR was performed using the inverse-variance weighted (IVW) method to test associations. The Jaccard index quantified the similarity of effects of each target across these outcomes, focusing on significant results after adjusting for multiple testing (FDR q < 0.05).
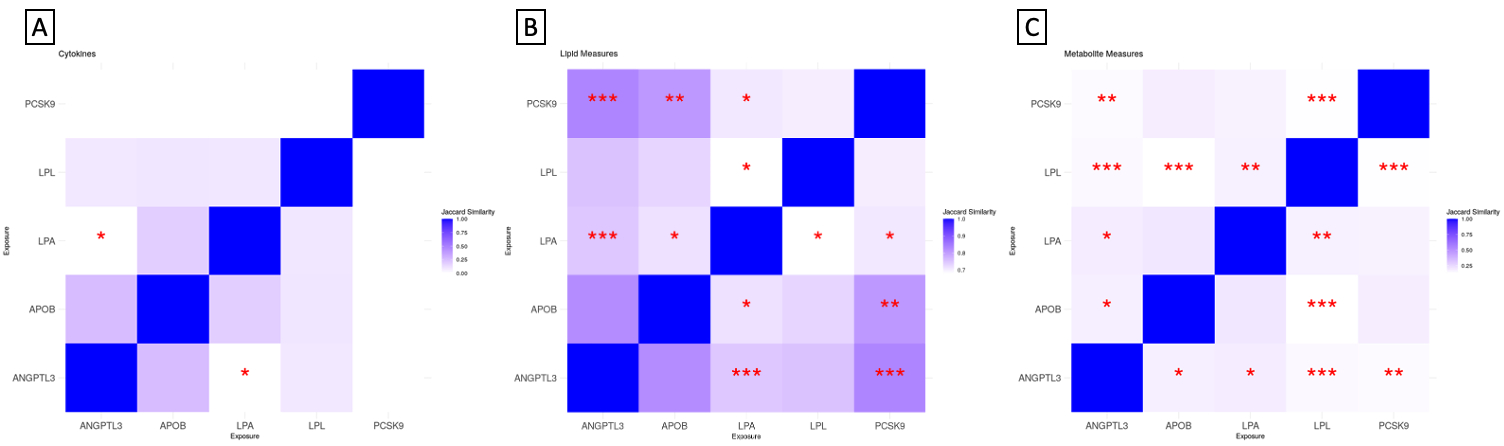
Results: The lipid-lowering targets were each significantly associated with between 1-17 cytokines, 184-220 lipid subfractions, and 307-396 metabolites (FDR q < 0.05). ANGPTL3 and PCSK9 showed the highest similarity of effects on lipid subfractions (Jaccard Index: 0.83, p < 0.001). LPA, APOB, and ANGPTL3 were the only targets with significant cytokine effects, with low similarity scores potentially suggesting their inflammatory signatures are unique. ANGPTL3 and LPA had the highest similarity of effects on metabolites (Jaccard Index: 0.19, p < 0.05).
Conclusion: Our analyses indicate that lipid-lowering targets influence metabolic signatures through shared and unique mechanisms. Further exploration of how cytokine, lipid, and metabolic profiles are impacted may help in the development of more effective medications.
Methods: The study focused on protein targets of novel lipid-lowering medications: PCSK9, APOB, LPA, LPL, and ANGPTL3. Instruments for PCSK9, LPL, and ANGPTL3 were derived from the UK Biobank Pharma Proteomics Project (54,219 participants), and for LPA and APOB from the Pan-UK Biobank (335,796 and 418,505 participants). These exposures were tested against outcomes including 40 cytokines (meta-analysis of 74,783 individuals), 249 lipid-subfraction measures (118,461 UK Biobank participants), and 1,400 metabolite measures (8,299 participants in the Canadian Longitudinal Study on Aging). Univariable MR was performed using the inverse-variance weighted (IVW) method to test associations. The Jaccard index quantified the similarity of effects of each target across these outcomes, focusing on significant results after adjusting for multiple testing (FDR q < 0.05).
Results: The lipid-lowering targets were each significantly associated with between 1-17 cytokines, 184-220 lipid subfractions, and 307-396 metabolites (FDR q < 0.05). ANGPTL3 and PCSK9 showed the highest similarity of effects on lipid subfractions (Jaccard Index: 0.83, p < 0.001). LPA, APOB, and ANGPTL3 were the only targets with significant cytokine effects, with low similarity scores potentially suggesting their inflammatory signatures are unique. ANGPTL3 and LPA had the highest similarity of effects on metabolites (Jaccard Index: 0.19, p < 0.05).
Conclusion: Our analyses indicate that lipid-lowering targets influence metabolic signatures through shared and unique mechanisms. Further exploration of how cytokine, lipid, and metabolic profiles are impacted may help in the development of more effective medications.
More abstracts on this topic:
12,13-diHOME Attenuates Pro-Arrhythmic Effect of High-Fat Diet
Buck Benjamin, Baer Lisa, Deschenes Isabelle, Chinthalapudi Krishna, Gallego-perez Daniel, Stanford Kristin, Hund Thomas, Areiza Natalia, Xu Xianyao, Elliott Austin, Wan Xiaoping, Nassal Drew, Lane Cemantha, Nirengi Shinsuke, James Natasha Maria
A Loss-of-Function Missense Variant in ANGPTL3 Exerts Protective Effects Against Kidney Disease RiskZhang David, Ritchie Marylyn, Rader Daniel, Cuchel Marina