Final ID: 062
Disease Trajectories of Cardiometabolic Diseases and Depression: Transition Patterns, Multiomics Signatures, Prognosis and Prediction
Abstract Body: Objective
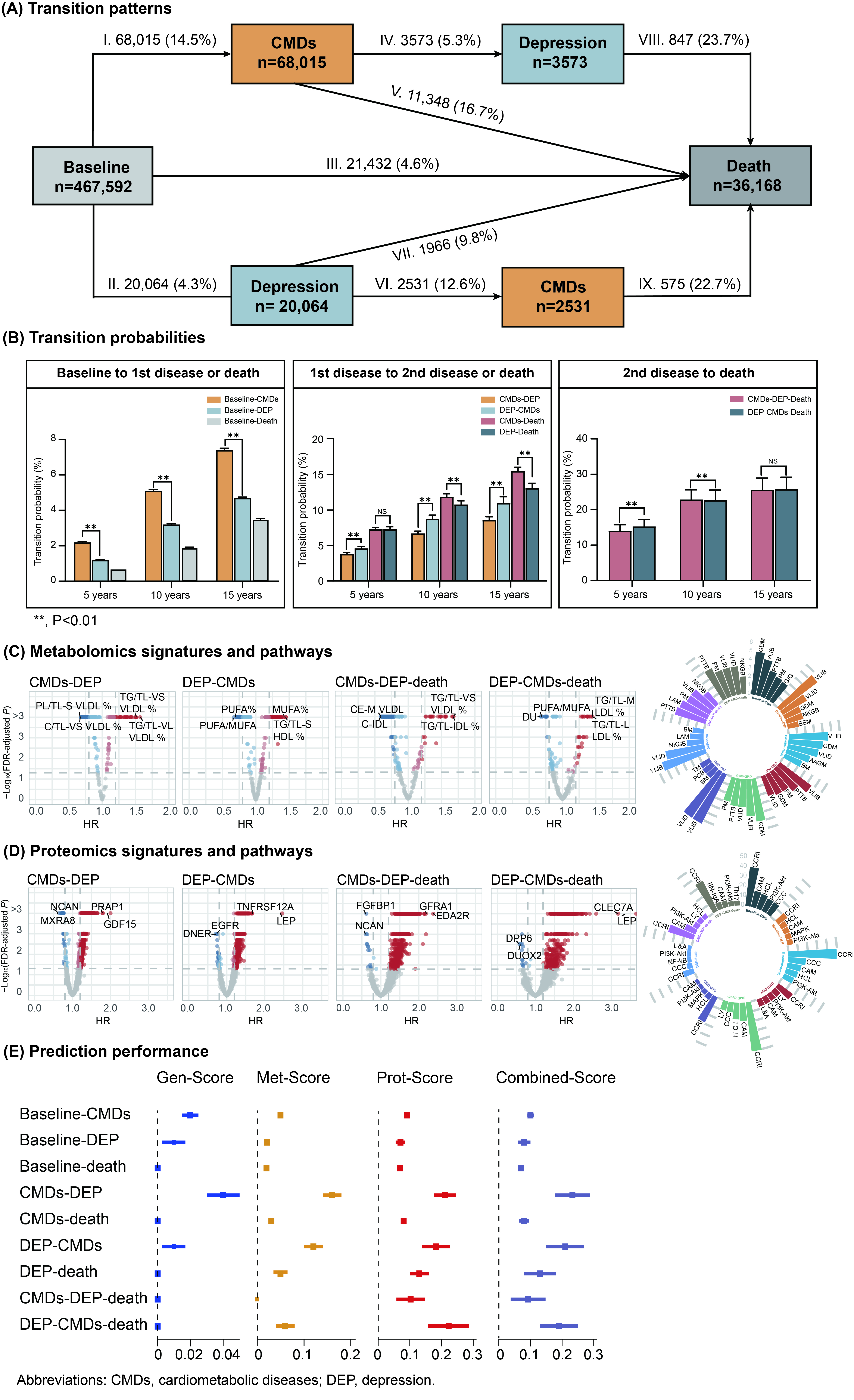
Cardiometabolic diseases (CMDs) and depression, among the most prevalent physical and mental diseases, frequently co-occur and are associated with a higher risk of premature mortality. However, the trajectories of their multimorbidity occurrence, underlying biological mechanisms, and early prediction remain poorly understood.
Methods
This study was conducted on 467,592 UK Biobank participants without baseline CMDs and depression. CMDs included type 2 diabetes, coronary artery disease, stroke, and heart failure. Multistate models were used to investigate transition probabilities and identify multiomics signatures for transitions from baseline to single morbidity and to multimorbidity. Multiomics prediction models were constructed using least absolute shrinkage and selection operator Cox regression. Model performance was evaluated by the change of Harrell's C statistic (Δ C-statistic) compared to the demographic model and area under receiver-operating characteristic curves (AUCs).
Results
During a median follow-up of 14.6 years, 64,442 participants developed CMDs alone, 17,533 developed depression alone, and 6104 developed multimorbidity. Depression preceding CMDs showed a 2.5% higher 15-year multimorbidity probability and 1.2% higher 5-year mortality risk than the reverse sequence. Multimorbidity was associated with a 14-26% higher mortality risk and 3.6-3.8 years shorter survival time compared to those without CMDs or depression. Distinct and shared multiomics signatures underlying disease trajectories were identified. Valine, leucine, and isoleucine biosynthesis and cytokine-cytokine receptor interaction pathways were implicated in both CMDs and depression progression. Proteomics scores showed superior prediction performance across nine disease transitions, with Δ C-statistic ranging from 0.07 for health-death to 0.22 for health-depression-CMDs-death, compared to the genomics (0-0.04) and metabolomics (0-0.16) scores. For 15-year outcome prediction, the proteomics scores model achieved AUCs of 0.65-0.87 for various transitions, significantly outperforming traditional risk factors-based and other omics models.
Conclusions
This study revealed distinct transition patterns, identified associated multiomics signatures, and constructed multiomics prediction models for the multimorbidity cluster of CMDs and depression.
Cardiometabolic diseases (CMDs) and depression, among the most prevalent physical and mental diseases, frequently co-occur and are associated with a higher risk of premature mortality. However, the trajectories of their multimorbidity occurrence, underlying biological mechanisms, and early prediction remain poorly understood.
Methods
This study was conducted on 467,592 UK Biobank participants without baseline CMDs and depression. CMDs included type 2 diabetes, coronary artery disease, stroke, and heart failure. Multistate models were used to investigate transition probabilities and identify multiomics signatures for transitions from baseline to single morbidity and to multimorbidity. Multiomics prediction models were constructed using least absolute shrinkage and selection operator Cox regression. Model performance was evaluated by the change of Harrell's C statistic (Δ C-statistic) compared to the demographic model and area under receiver-operating characteristic curves (AUCs).
Results
During a median follow-up of 14.6 years, 64,442 participants developed CMDs alone, 17,533 developed depression alone, and 6104 developed multimorbidity. Depression preceding CMDs showed a 2.5% higher 15-year multimorbidity probability and 1.2% higher 5-year mortality risk than the reverse sequence. Multimorbidity was associated with a 14-26% higher mortality risk and 3.6-3.8 years shorter survival time compared to those without CMDs or depression. Distinct and shared multiomics signatures underlying disease trajectories were identified. Valine, leucine, and isoleucine biosynthesis and cytokine-cytokine receptor interaction pathways were implicated in both CMDs and depression progression. Proteomics scores showed superior prediction performance across nine disease transitions, with Δ C-statistic ranging from 0.07 for health-death to 0.22 for health-depression-CMDs-death, compared to the genomics (0-0.04) and metabolomics (0-0.16) scores. For 15-year outcome prediction, the proteomics scores model achieved AUCs of 0.65-0.87 for various transitions, significantly outperforming traditional risk factors-based and other omics models.
Conclusions
This study revealed distinct transition patterns, identified associated multiomics signatures, and constructed multiomics prediction models for the multimorbidity cluster of CMDs and depression.
More abstracts on this topic:
A machine learning model for individualized risk prediction of ischemic heart disease in people with hypertension in Thailand
Sakboonyarat Boonsub, Poovieng Jaturon, Rangsin Ram
Association of Eicosanoid Metabolites with Insulin ResistanceParekh Juhi, Chitsazan Mandana, Roshandelpoor Athar, Alotaibi Mona, Jain Mohit, Cheng Susan, Ho Jennifer