Final ID: MP1117
Decoding the Regulatory Landscape of KMT2D in Heart Development
Abstract Body (Do not enter title and authors here): Background: Hypoplastic Left Heart Syndrome (HLHS) remains the most lethal form of congenital heart disease (CHD). The next generation of therapies are predicated on understanding the molecular bases for HLHS. We created kmt2d null zebrafish that recapitulate features of human Kabuki Syndrome (KS), including hypoplastic heart and death from circulatory collapse. Notch signaling is hyperactivated in endocardial cells, with aberrant endocardial-to-mesenchymal transition (EMT) that occludes the ventricle; an effect that is rescued by Notch inhibition. Notch hyperactivation and aberrant EMT are hallmarks of endocardial fibroelastosis (EFE) observed in HLHS.
Objective: Define how KMT2D deletion disrupts the epigenetic landscape controlling Notch signaling in zebrafish heart development and human induced pluripotent stem cell (iPSC)-derived cardiac lineages.
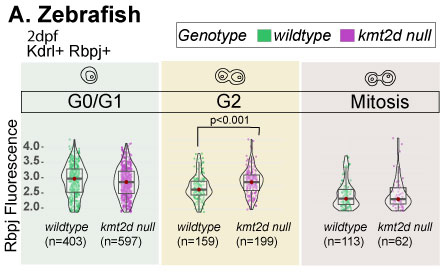
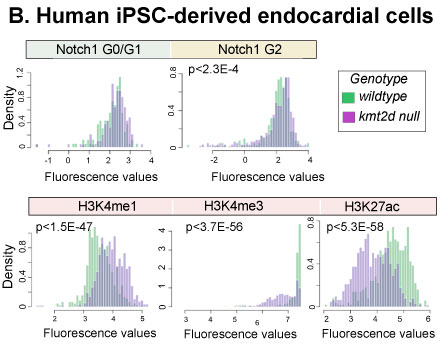
Methods: We performed immunofluorescence followed by Fluorescence Activated Cell Sorting (FACS) on Tg(kdrl:GFP) WT and kmt2d-null embryos at 2 dpf (n=3 each group), gating on kdrl+ (endocardial) cells, measuring Rbpj fluorescence as a marker of Notch activation and DAPI for cell cycle analysis. Single-cell histone profiling was performed on WT and KMT2D null human iPSC-derived endocardial cells (n=3 each group), using fluorescently labelled antibodies against H3K4me1, H3K4me3, H3K27ac, H3K27me3, Notch1 and cell-cycle proteins.
Results: Notch signaling was preferentially elevated in G2 cell-cycle endocardial cells in kmt2d-null embryos, prior to development of heart hypoplasia (p<0.001, n=62-597 cells each group). KMT2D null human iPSC-derived endocardial cells phenocopied the zebrafish null mutants, with increased Notch1 specifically in G2 cell-cycle state compared to WT (p<2.3E-4). H3K4me1 was increased (p <1.5E-47), while H3K4me3 (p<3.7E-56) and H3K27ac (p<5.3E-58) signals were decreased in KMT2D null endocardial cells (n=529-665 cells each group), indicating a proportion of cells that are poised for activation, but transcriptionally silent.
Conclusion: KMT2D functions to control Notch expression in endocardial cells in G2 cell cycle. KMT2D deletion disrupts chromatin states that usually repress Notch signaling in endocardial cells, triggering excessive Notch and aberrant EMT. Ongoing experiments are defining the gene regulatory networks modulated by KMT2D that contribute to heart chamber formation. Our findings are relevant beyond KS alone, by furthering our understanding of EFE pathogenesis in HLHS.
Objective: Define how KMT2D deletion disrupts the epigenetic landscape controlling Notch signaling in zebrafish heart development and human induced pluripotent stem cell (iPSC)-derived cardiac lineages.
Methods: We performed immunofluorescence followed by Fluorescence Activated Cell Sorting (FACS) on Tg(kdrl:GFP) WT and kmt2d-null embryos at 2 dpf (n=3 each group), gating on kdrl+ (endocardial) cells, measuring Rbpj fluorescence as a marker of Notch activation and DAPI for cell cycle analysis. Single-cell histone profiling was performed on WT and KMT2D null human iPSC-derived endocardial cells (n=3 each group), using fluorescently labelled antibodies against H3K4me1, H3K4me3, H3K27ac, H3K27me3, Notch1 and cell-cycle proteins.
Results: Notch signaling was preferentially elevated in G2 cell-cycle endocardial cells in kmt2d-null embryos, prior to development of heart hypoplasia (p<0.001, n=62-597 cells each group). KMT2D null human iPSC-derived endocardial cells phenocopied the zebrafish null mutants, with increased Notch1 specifically in G2 cell-cycle state compared to WT (p<2.3E-4). H3K4me1 was increased (p <1.5E-47), while H3K4me3 (p<3.7E-56) and H3K27ac (p<5.3E-58) signals were decreased in KMT2D null endocardial cells (n=529-665 cells each group), indicating a proportion of cells that are poised for activation, but transcriptionally silent.
Conclusion: KMT2D functions to control Notch expression in endocardial cells in G2 cell cycle. KMT2D deletion disrupts chromatin states that usually repress Notch signaling in endocardial cells, triggering excessive Notch and aberrant EMT. Ongoing experiments are defining the gene regulatory networks modulated by KMT2D that contribute to heart chamber formation. Our findings are relevant beyond KS alone, by furthering our understanding of EFE pathogenesis in HLHS.
More abstracts on this topic:
Adult Congenital Heart Disease and Atrial Fibrillation: A Double Burden on Cardiovascular Outcomes
Menassa Yara, Nahle Tarek, Lim Chanho, Hassan Abboud, Liu Yingshuo, Assaf Ala', El Hajjar Abdel Hadi, Noujaim Charbel, Dagher Lilas, Mekhael Mario, Rao Swati, Bsoul Mayana, Kreidieh Omar, Pandey Amitabh, Borgi Jamil, Marrouche Nassir, Atasi Mohammad Montaser, Jia Yishi, Feng Han, Abou Khalil Michel, Massad Christian, Bidaoui Ghassan, Younes Hadi
Common and Divergent Cellular Etiologies Underlying Hypoplastic Left Heart Syndrome and Hypoplastic Right Heart SyndromeYu Yang, Wang Cankun, Ye Shiqiao, Qin Hannah, Mcnutt Megan, Alonzo Matthew, Texter Karen, Garg Vidu, Zhao Mingtao