Final ID: Mo4047
Towards More Sensitive Detection of Cardiovascular Proteolytic Signals: A Substrate Phage-Display Based Approach
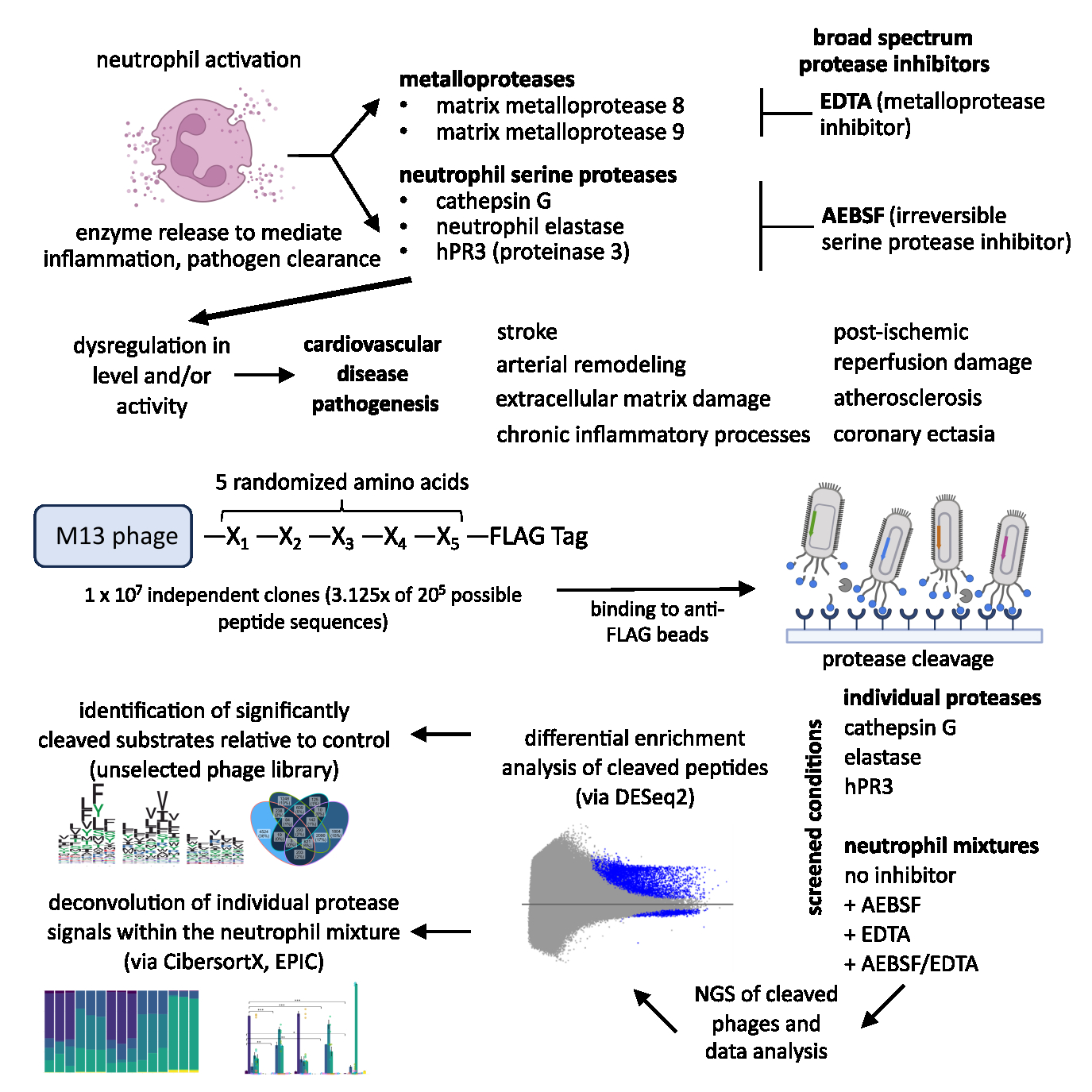
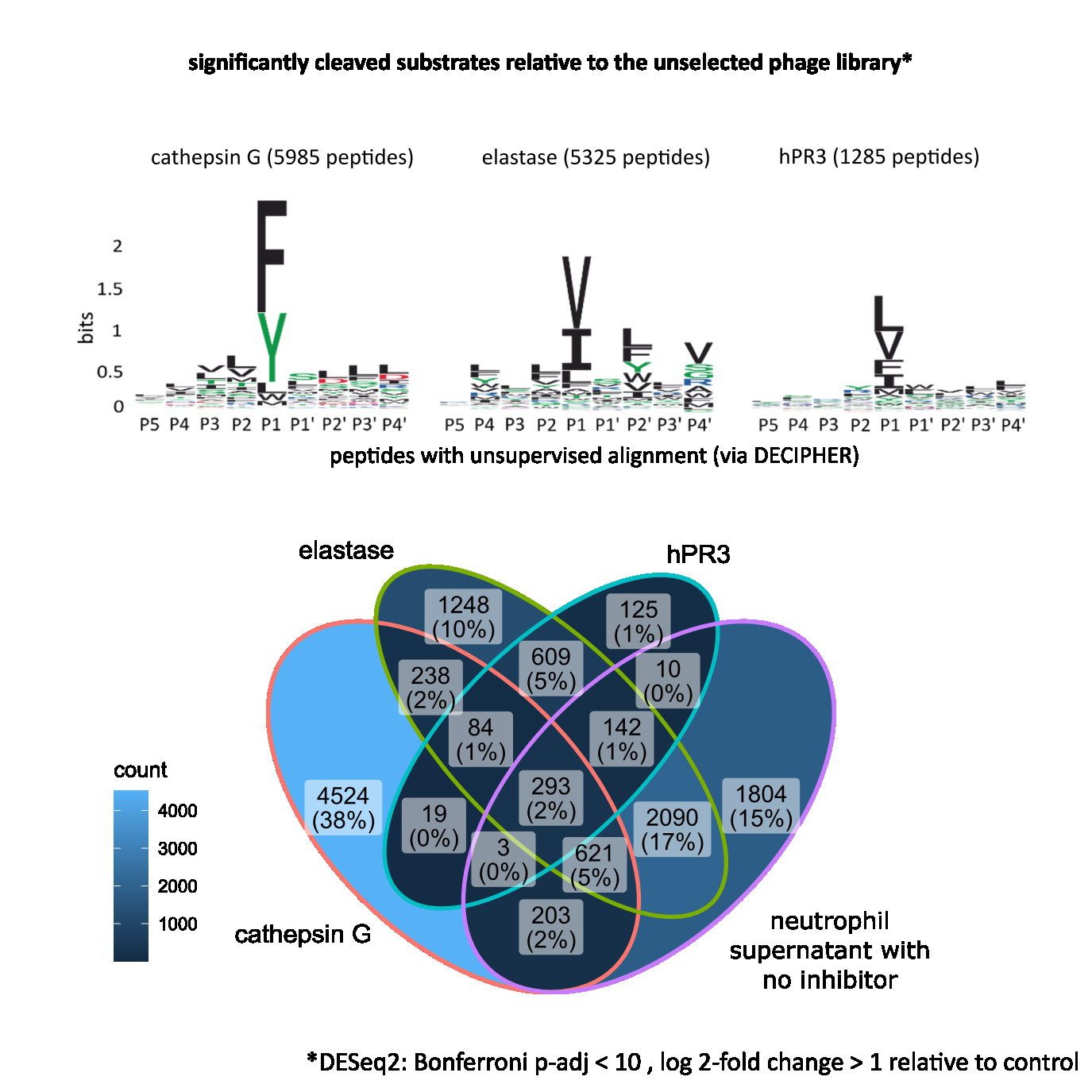
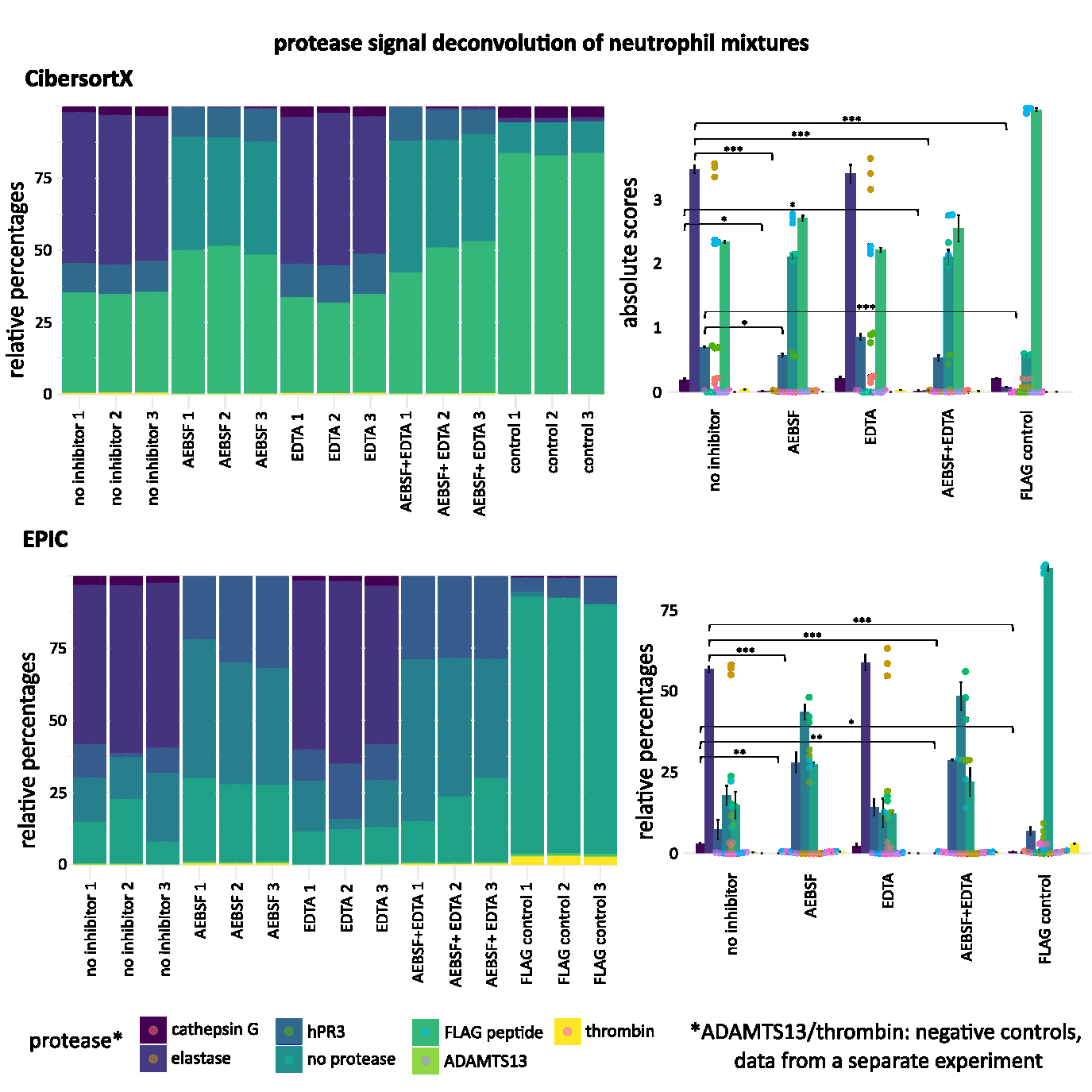
Abstract Body (Do not enter title and authors here): Introduction: Dysregulated protease activity is a key early contributor to cardiovascular diseases, with neutrophil serine proteases (NSPs) implicated in the pathogenesis of stroke, chronic inflammation, atherosclerosis, and coronary ectasia. However, detecting protease activity in complex biological mixtures is limited, because current substrate-based probes may lack sufficient specificity and require prior knowledge of protease content to select probes. Objective: Our study aims to develop Deep Protease Profiling as a high-resolution method for the specific and functional characterization of protease activity in mixtures and validate its use with activated neutrophil supernatants. Methods: A random 5 amino acid substrate phage display library was constructed and screened against 3 NSPs: elastase, cathepsin G (CSG), and human proteinase 3 (hPR3). The library was also screened against the supernatant of phorbol myristate acetate (PMA)-activated neutrophils alone or in the presence of protease inhibitors. Each condition was conducted in triplicate. Cleaved phages were isolated, and cleaved sequences were identified by high-throughput sequencing. Sequencing data were analyzed by adapting algorithms used for RNAseq applications. Results: Substrate specificities for each NSP aligned with existing literature, prior protease screens, and known active site architecture. Proteolysis detected in the activated neutrophil supernatant was attributable to elastase (62%), CSG (22%), and hPR3 (3%). Peptides identified from the purified NSP reactions captured 66% of total activity in the neutrophil supernatants. Deconvolution algorithms objectively captured substrate profiles of >1M peptides to quantify protease activity across neutrophil mixtures, with significantly reduced elastase, CSG, and hPR3 activity identified in neutrophil supernatants treated with AEBSF. Results across two algorithms tested (CibersortX, EPIC) were highly congruent. We further found unique substrates distinct for each protease and classified physiologically relevant cleavage motifs, with validation by MEROPs databases. Conclusion: We demonstrated a global, unbiased, and systematic method of unprecedented breadth to detect and deconvolute protease activity in complex biological mixtures. Future studies will expand Deep Protease Profiling to other proteases, mixtures, and clinical samples, with promise as an emerging platform for developing novel diagnostic tools to manage disease in patients.
More abstracts on this topic:
AI-Derived Retinal Vasculature Features Predict Cardiovascular Risk in Patients with Chronic Kidney Disease: Insights from the CRIC Study
Dhamdhere Rohan, Modanwal Gourav, Rahman Mahboob, Al-kindi Sadeer, Madabhushi Anant
ADP-Ribosylation In a Mouse Model of Atherosclerosis: a Potential Novel Link Between Dyslipidemia and Inflammation in Cardiovascular DiseaseDelwarde Constance, Mlynarchik Andrew, Perez Katelyn, Campedelli Alesandra, Sonawane Abhijeet, Aikawa Elena, Singh Sasha, Aikawa Masanori, Santinelli Pestana Diego, Kasai Taku, Kuraoka Shiori, Nakamura Yuto, Okada Takeshi, Decano Julius, Chelvanambi Sarvesh, Ge Rile