Final ID: MDP1295
The plasma cell-free transcriptome non-invasively profiles the immune response and myocardial damage in immune checkpoint inhibitor-induced myocarditis.
Abstract Body (Do not enter title and authors here): Introduction
Immune checkpoint inhibitor-induced myocarditis (ICI-m) is a severe complication of cancer immunotherapy, marked by cardiac inflammation. Despite its high mortality rate, diagnosing ICI-m remains challenging due to the lack of sensitive and non-invasive diagnostic modalities. Plasma cell-free mRNA (cf-mRNA) has recently been highlighted as a promising tool for non-invasive tissue profiling.
Hypothesis
The plasma cf-mRNA profile contains disease-specific immune and tissue transcriptomic signatures of ICI-m that can be leveraged for disease-specific precision diagnostics.
Aims
To establish and validate a novel cf-mRNA bench and bioinformatic pipeline for noninvasive profiling of the immune and tissue landscape of patients with ICI-m.
Methods
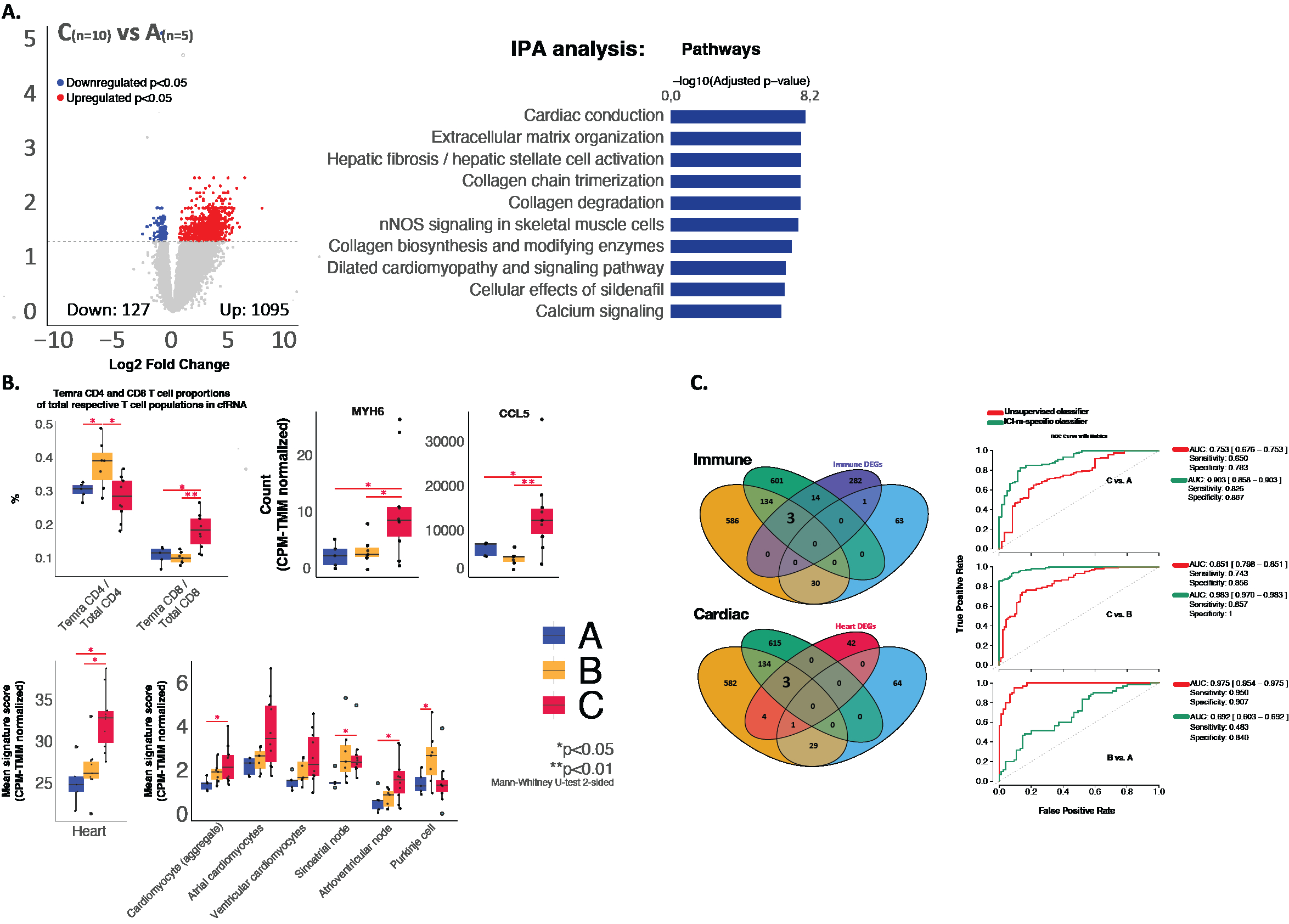
We isolated, DNase-treated, and sequenced bulk cfRNA from frozen plasma of patients with ICI-treated cancer patients with no autoimmunity (group A, n=5), extracardiac autoimmunity (group B, n=7), and ICI-m with or without extracardiac autoimmunity (group C, n=10). Publicly available healthy control cfRNA was downloaded (n=30) as external controls. We used cell type-specific gene panels curated from single-cell RNA-sequencing datasets of circulating immune cells and cardiac cells for cellular deconvolution of the cf-mRNA. Signature scores of the sum of the counts of each cell type-specific gene were used to compare cell type contributions between patient groups.
Results
Cancer and inflammatory gene pathways were significantly enriched in the cf-mRNA of all ICI-treated cancer patients. Cardiac pathways and conduction genes were significantly enriched in group C vs. A and B patients. Signature scores of Temra CD8+ T cells and cardiomyocytes, and gene expression of CCL5 and MYH6 were elevated in group C patients. Our custom ICI-m classifier containing 3 cardiac- and 3 immune cell-specific genes differentiated group C from A (AUC 0.903, 95% CI 0.858-0.903) and B patients (AUC 0.983, 95% CI 0.970-0.983) effectively, outperforming a diagnostic classifier obtained from unsupervised feature selection from all differentially expressed genes.
Conclusions
Using our novel plasma cf-mRNA platform, we developed a diagnostic classifier that captures the disease-specific immune and tissue transcriptomic signatures of ICI-m, highlighting the advantages of this disease-specific approach over conventional biomarker discovery, and demonstrating the broader implications of the platform for the field of precision diagnostics.
Immune checkpoint inhibitor-induced myocarditis (ICI-m) is a severe complication of cancer immunotherapy, marked by cardiac inflammation. Despite its high mortality rate, diagnosing ICI-m remains challenging due to the lack of sensitive and non-invasive diagnostic modalities. Plasma cell-free mRNA (cf-mRNA) has recently been highlighted as a promising tool for non-invasive tissue profiling.
Hypothesis
The plasma cf-mRNA profile contains disease-specific immune and tissue transcriptomic signatures of ICI-m that can be leveraged for disease-specific precision diagnostics.
Aims
To establish and validate a novel cf-mRNA bench and bioinformatic pipeline for noninvasive profiling of the immune and tissue landscape of patients with ICI-m.
Methods
We isolated, DNase-treated, and sequenced bulk cfRNA from frozen plasma of patients with ICI-treated cancer patients with no autoimmunity (group A, n=5), extracardiac autoimmunity (group B, n=7), and ICI-m with or without extracardiac autoimmunity (group C, n=10). Publicly available healthy control cfRNA was downloaded (n=30) as external controls. We used cell type-specific gene panels curated from single-cell RNA-sequencing datasets of circulating immune cells and cardiac cells for cellular deconvolution of the cf-mRNA. Signature scores of the sum of the counts of each cell type-specific gene were used to compare cell type contributions between patient groups.
Results
Cancer and inflammatory gene pathways were significantly enriched in the cf-mRNA of all ICI-treated cancer patients. Cardiac pathways and conduction genes were significantly enriched in group C vs. A and B patients. Signature scores of Temra CD8+ T cells and cardiomyocytes, and gene expression of CCL5 and MYH6 were elevated in group C patients. Our custom ICI-m classifier containing 3 cardiac- and 3 immune cell-specific genes differentiated group C from A (AUC 0.903, 95% CI 0.858-0.903) and B patients (AUC 0.983, 95% CI 0.970-0.983) effectively, outperforming a diagnostic classifier obtained from unsupervised feature selection from all differentially expressed genes.
Conclusions
Using our novel plasma cf-mRNA platform, we developed a diagnostic classifier that captures the disease-specific immune and tissue transcriptomic signatures of ICI-m, highlighting the advantages of this disease-specific approach over conventional biomarker discovery, and demonstrating the broader implications of the platform for the field of precision diagnostics.
More abstracts on this topic:
A Multi-Population-First Approach Leveraging UK Biobank (UKBB) and All of Us (AoU) Datasets Reveals Higher Cardiomyopathy Variant Burden in Individuals with Myocarditis
Gurumoorthi Manasa, Khanji Mohammed, Munroe Patricia, Petersen Steffen, Landstrom Andrew, Chahal Anwar, Hesse Kerrick, Asatryan Babken, Shah Ravi, Sharaf Dabbagh Ghaith, Wolfe Rachel, Shyam Sundar Vijay, Mohiddin Saidi, Aung Nay
90-Day Readmission Rates, Predictors, and Causes of Readmission After Placement of Left Atrial Appendage Occlusion Device in Patients With history of different malignancies: National Readmission Database analysisQuevedo Ramirez Andres, Teaima Taha, Jha Vivek, Ibarra Joshua, Soon-shiong Raquel, Gomez Valencia Javier