Final ID: 4119052
Redefining the phenotypic continuum of cardiovascular disease using machine learning
Abstract Body (Do not enter title and authors here): Background
Current approaches that define cardiovascular disease rely on assignment of traditional diagnostic labels which may not reflect the natural continuum of phenotypic expression, influenced by genetic and environmental modifiers, or adequately identify groups with shared molecular mechanisms. Machine learning can leverage advances in genetic and imaging characterisation to reclassify phenotypic diversity along a continuum, from health to disease.
Research Aims
Our study aims to build a tree-like classification of cardiovascular phenotypes in the community where branches represent subjects with shared features, ordered by their severity. Using dilated cardiomyopathy (DCM) and hypertrophic cardiomyopathy (HCM) as exemplar conditions with opposing phenotypes, we aim to demonstrate the interaction between genetic and environmental modifiers of disease expression.
Methods
We analysed cardiovascular magnetic resonance (CMR) imaging, electrocardiogram (ECG), biomarkers and clinical data from participants in the UK Biobank. Using unsupervised learning of multiparametric data, we built a tree of phenotypic expression. We projected the risk of DCM and HCM cases, and associated rare and common variant risk, onto the tree structure.
Results
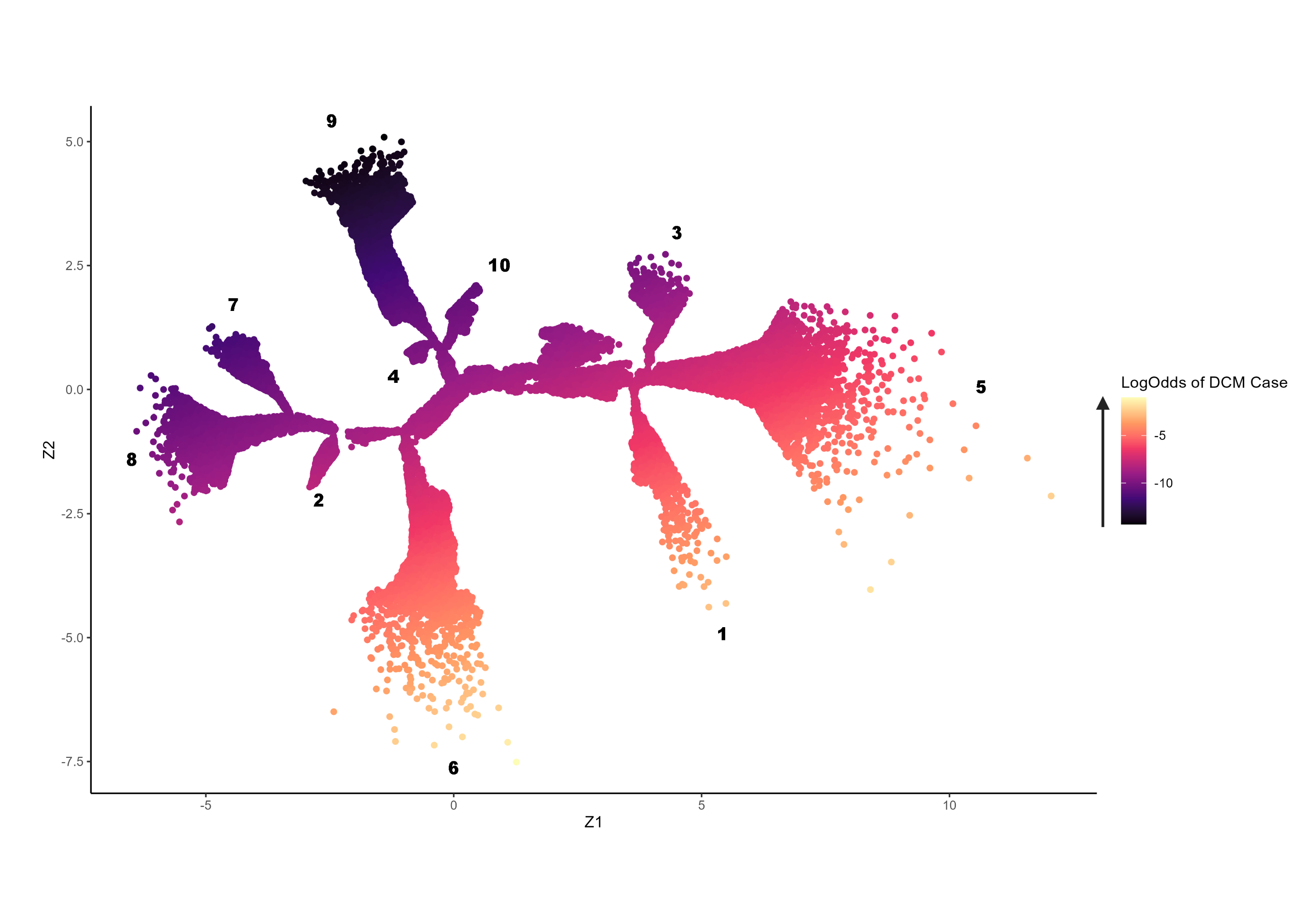
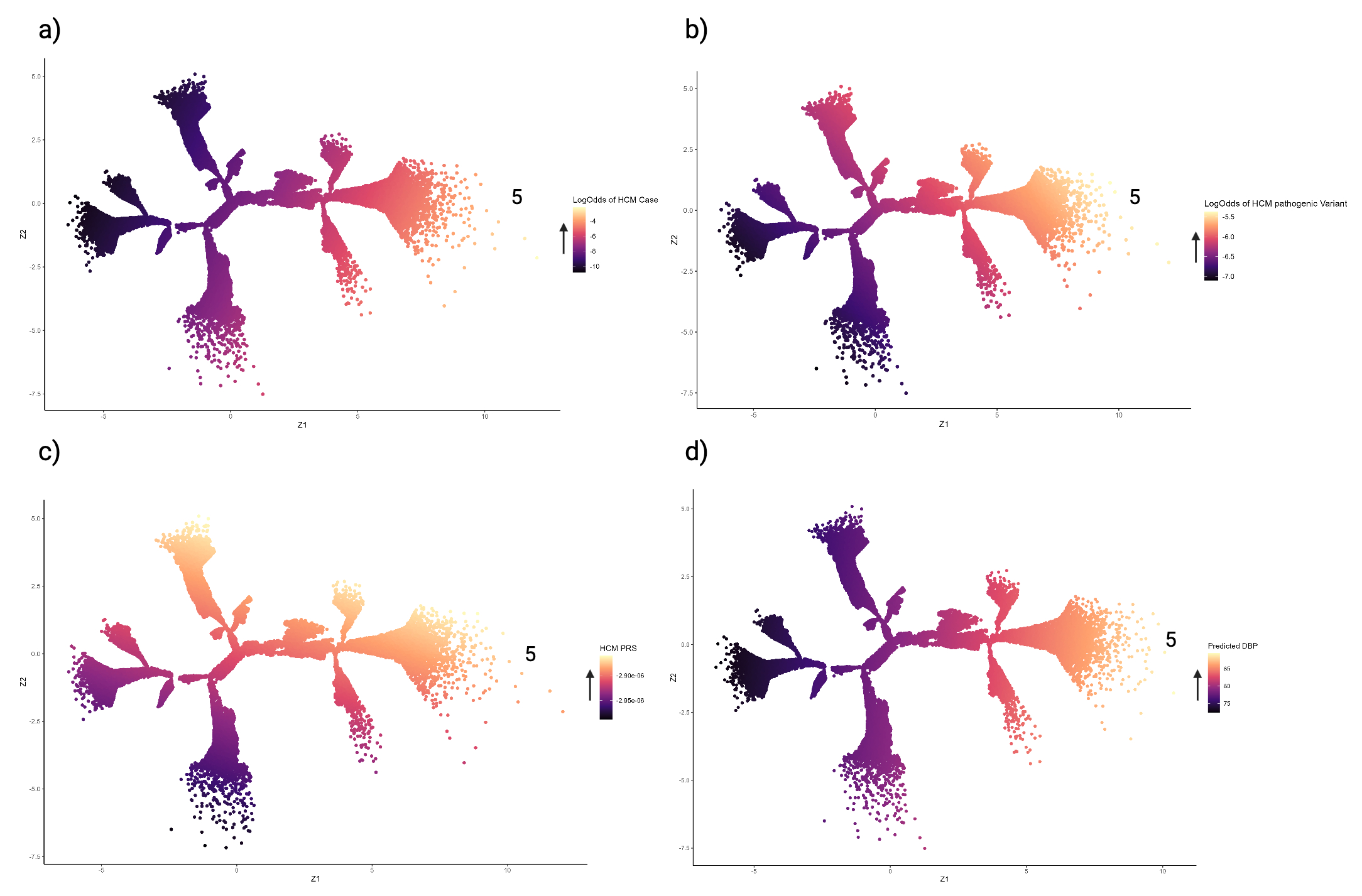
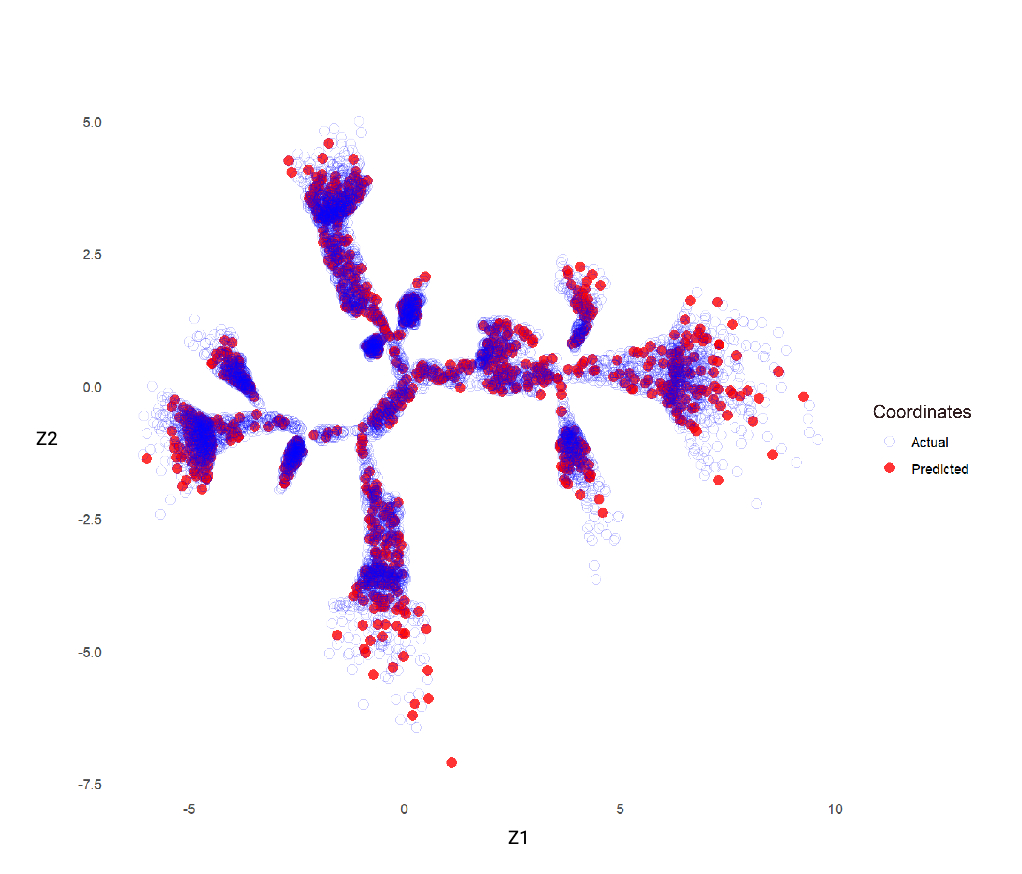
Ten main branches were discovered, using data from 41,525 participants (51.7 % female, median age 65 yrs [IQR: 58, 70]). The extremities of branches 1 and 6 were enriched for DCM cases (p <0.05) and participants with reduced ejection fraction and high left ventricular volumes (p <0.05). Compared with subjects at the tree centre, distal participants in branch 1 had increased blood pressure (BP) and body mass index (BMI), whilst those in branch 6 were more likely to have high HbA1c levels and carry a pathogenic DCM variant (p <0.05). The extremity of branch 5 was enriched for HCM cases, rare sarcomeric variants (p = 0.004) and high polygenic risk (p <0.001). Whilst the ends of branches 3 and 9 had high polygenic risk for HCM, they were less likely to have elevated BP and BMI, and had reduced likelihood of HCM expression, compared with branch 5. A phenomapping model was trained to accurately project unseen subjects onto the tree (R2 0.98).
Conclusions
We present a tree-like continuum of cardiovascular phenotypes, providing a novel framework for mechanistic discovery and exploration of genotype-phenotype associations. Phenomapping of unseen subjects enables personalised estimation of genetic and cardiovascular risk.
Current approaches that define cardiovascular disease rely on assignment of traditional diagnostic labels which may not reflect the natural continuum of phenotypic expression, influenced by genetic and environmental modifiers, or adequately identify groups with shared molecular mechanisms. Machine learning can leverage advances in genetic and imaging characterisation to reclassify phenotypic diversity along a continuum, from health to disease.
Research Aims
Our study aims to build a tree-like classification of cardiovascular phenotypes in the community where branches represent subjects with shared features, ordered by their severity. Using dilated cardiomyopathy (DCM) and hypertrophic cardiomyopathy (HCM) as exemplar conditions with opposing phenotypes, we aim to demonstrate the interaction between genetic and environmental modifiers of disease expression.
Methods
We analysed cardiovascular magnetic resonance (CMR) imaging, electrocardiogram (ECG), biomarkers and clinical data from participants in the UK Biobank. Using unsupervised learning of multiparametric data, we built a tree of phenotypic expression. We projected the risk of DCM and HCM cases, and associated rare and common variant risk, onto the tree structure.
Results
Ten main branches were discovered, using data from 41,525 participants (51.7 % female, median age 65 yrs [IQR: 58, 70]). The extremities of branches 1 and 6 were enriched for DCM cases (p <0.05) and participants with reduced ejection fraction and high left ventricular volumes (p <0.05). Compared with subjects at the tree centre, distal participants in branch 1 had increased blood pressure (BP) and body mass index (BMI), whilst those in branch 6 were more likely to have high HbA1c levels and carry a pathogenic DCM variant (p <0.05). The extremity of branch 5 was enriched for HCM cases, rare sarcomeric variants (p = 0.004) and high polygenic risk (p <0.001). Whilst the ends of branches 3 and 9 had high polygenic risk for HCM, they were less likely to have elevated BP and BMI, and had reduced likelihood of HCM expression, compared with branch 5. A phenomapping model was trained to accurately project unseen subjects onto the tree (R2 0.98).
Conclusions
We present a tree-like continuum of cardiovascular phenotypes, providing a novel framework for mechanistic discovery and exploration of genotype-phenotype associations. Phenomapping of unseen subjects enables personalised estimation of genetic and cardiovascular risk.
More abstracts on this topic:
A Case of Concomitant Wild-Type Transthyretin and Systemic Light Chain Amyloidosis Involving Separate Organs
Chiu Leonard, Afrough Aimaz, Nadeem Urooba, Jebakumar Deborah, Grodin Justin
A Case of Hypertrophic Cardimyopathy: Digenic Variants of Uncertain Significance Mutations in MHY7 and RYR2 GenesDurukan Selina, Uzunoglu Ekin, Farahmandsadr Maryam, Soffer Daniel