Final ID: We0005
Spatial Transcriptomic Analysis of Human Tibial Vessels Reveals Bilayered Media
Abstract Body: Introduction
The arterial wall is formed of three layers—intima, media, and adventitia—from inside to out. However, dissection cleavage planes occur within the media, where we expect no natural tissue plane. Dissections can lead to clinical complications such as aneurysmal dilation, rupture or occlusion in nearly any artery. The mechanisms driving dissection remain poorly understood, and our clinical terminology is even inaccurate, with the cleaved layer referred to as an “intimal flap”. Whether medial layer architecture can promote dissection is unknown, but spatial transcriptomic technologies, which allow cellular-level gene expression mapping in intact tissue, can provide unique cell and molecular level insight into transmural architecture.
Hypothesis
We predict spatial transcriptomic analysis will elucidate cell and molecular organization within the arterial media, providing insight into the pathogenesis of arterial dissection in human vascular specimens.
Methods
Using a non-diseased human tibial vascular specimen collected from the Mid-America Transplant Center in St. Louis, MO, we applied the Xenium platform (10X Genomics) with a custom 480 gene panel. After quality control filtering low-quality cells and normalization, we applied k-means clustering to identify cell-type clusters. To render the transmural location, we developed a vessel coordinate system. We calculated the center point of the vessel for the origin, and depth was computed as distance from origin to cell, normalized by distance from origin to vessel boundary. Cell locations were mapped by transmural location.
Results
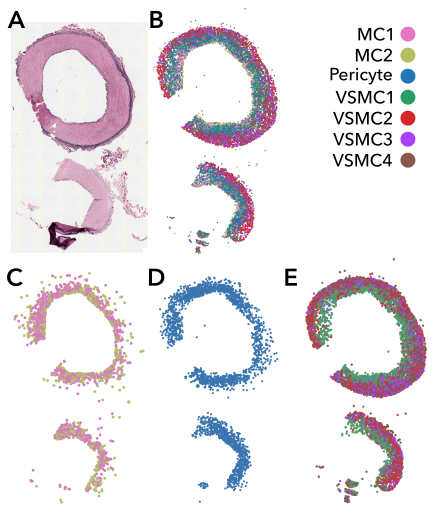
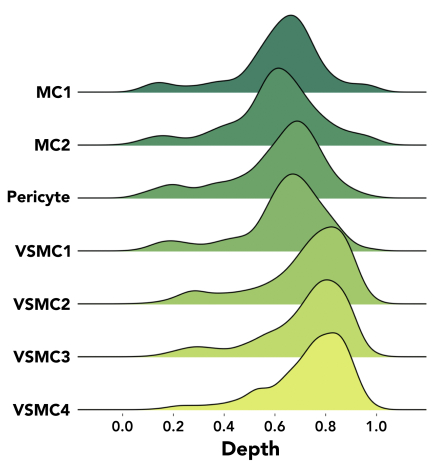
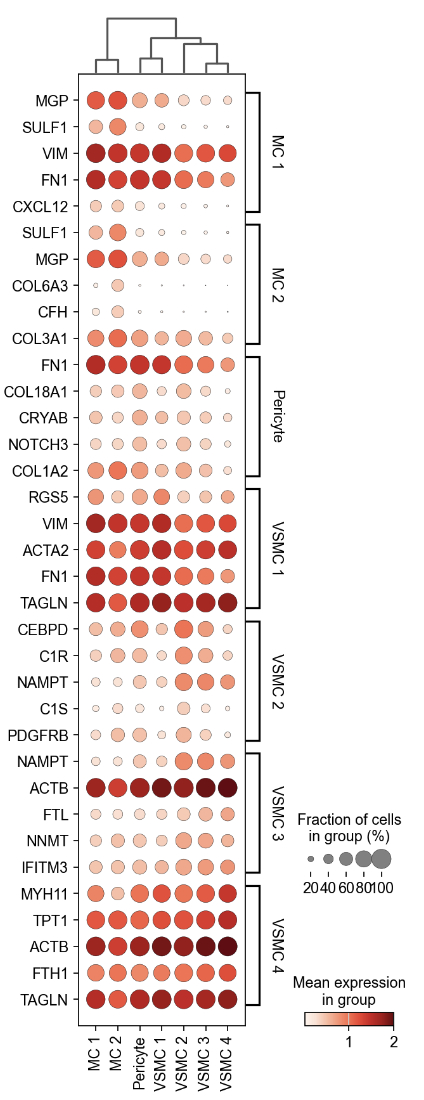
Using clustering techniques we annotated 10 cell types, including 1 endothelial cell, 1 fibroblast, 1 macrophage, 2 mesenchymal cell (MC), 1 pericyte and 4 vascular smooth muscle cell (VSMC) niches. Excluding endothelial and adventitial cell clusters, we visualized a bilayered appearance to the media (Figure 1). Cell depth was computed and displayed as a ridgeline plot (Figure 2). We also examined expression levels of the 5 most cluster-enriched genes by cell type, which suggest unique layer markers. (Figure 3).
Conclusions
With spatial transcriptomics, we identified two distinct cellular layers of the media with unique gene expression, suggesting a potential natural cleavage plane that may be important for dissection pathology. While this study evaluates non-diseased vascular architecture, we anticipate greater insights by applying this technology to dissected vascular tissues.
The arterial wall is formed of three layers—intima, media, and adventitia—from inside to out. However, dissection cleavage planes occur within the media, where we expect no natural tissue plane. Dissections can lead to clinical complications such as aneurysmal dilation, rupture or occlusion in nearly any artery. The mechanisms driving dissection remain poorly understood, and our clinical terminology is even inaccurate, with the cleaved layer referred to as an “intimal flap”. Whether medial layer architecture can promote dissection is unknown, but spatial transcriptomic technologies, which allow cellular-level gene expression mapping in intact tissue, can provide unique cell and molecular level insight into transmural architecture.
Hypothesis
We predict spatial transcriptomic analysis will elucidate cell and molecular organization within the arterial media, providing insight into the pathogenesis of arterial dissection in human vascular specimens.
Methods
Using a non-diseased human tibial vascular specimen collected from the Mid-America Transplant Center in St. Louis, MO, we applied the Xenium platform (10X Genomics) with a custom 480 gene panel. After quality control filtering low-quality cells and normalization, we applied k-means clustering to identify cell-type clusters. To render the transmural location, we developed a vessel coordinate system. We calculated the center point of the vessel for the origin, and depth was computed as distance from origin to cell, normalized by distance from origin to vessel boundary. Cell locations were mapped by transmural location.
Results
Using clustering techniques we annotated 10 cell types, including 1 endothelial cell, 1 fibroblast, 1 macrophage, 2 mesenchymal cell (MC), 1 pericyte and 4 vascular smooth muscle cell (VSMC) niches. Excluding endothelial and adventitial cell clusters, we visualized a bilayered appearance to the media (Figure 1). Cell depth was computed and displayed as a ridgeline plot (Figure 2). We also examined expression levels of the 5 most cluster-enriched genes by cell type, which suggest unique layer markers. (Figure 3).
Conclusions
With spatial transcriptomics, we identified two distinct cellular layers of the media with unique gene expression, suggesting a potential natural cleavage plane that may be important for dissection pathology. While this study evaluates non-diseased vascular architecture, we anticipate greater insights by applying this technology to dissected vascular tissues.
More abstracts on this topic:
Loss of Hsp90β Enhances Mitochondrial Cristae Density and Preserves Mitochondrial Structure Under Stress
Roberts Richard, Wisner Halley, Sen Sujoita, Lam Chi
Arterial Stiffness Mechanisms and Orthostatic Hypotension in The Systolic Blood Pressure Intervention TrialGepner Adam, Jaeger Byron, Hughes Timothy, Upadhya Bharathi, Kitzman Dalane, Supiano Mark, Pewowaruk Ryan