Final ID: 045
Genome-wide Association Study for Metabolites in the Tryptophan Metabolism Pathway among African American Patients with Chronic Kidney Disease
Abstract Body: Introduction: Impaired tryptophan metabolism is a signature in chronic kidney disease (CKD), leading to adverse cardiovascular outcomes. Genome-wide association studies (GWAS) in CKD patients may uncover genes involved in tryptophan metabolism that are only detectable in individuals with CKD. Therefore, we conducted GWAS for tryptophan related metabolites among 1,322 African American participants of the Chronic Renal Insufficiency Cohort (CRIC), a minority patient group that genomic studies of tryptophan are lacking.
Methods: Thirty-two tryptophan-related metabolites were profiled from baseline 24-hour urine samples. Genome-wide array data were imputed to the Trans-Omics for Precision Medicine reference panel. After stringent quality control, GWAS of each metabolite was conducted, adjusting for age, sex, and 10 genetic principal components. Variants with P < 5×10-8 were considered significant. Conditional association analysis using GCTA-cojo, followed by fine-mapping using SuSiE, was performed on the 1000 kb region around a significant variant to identify independent signals and causal variants.
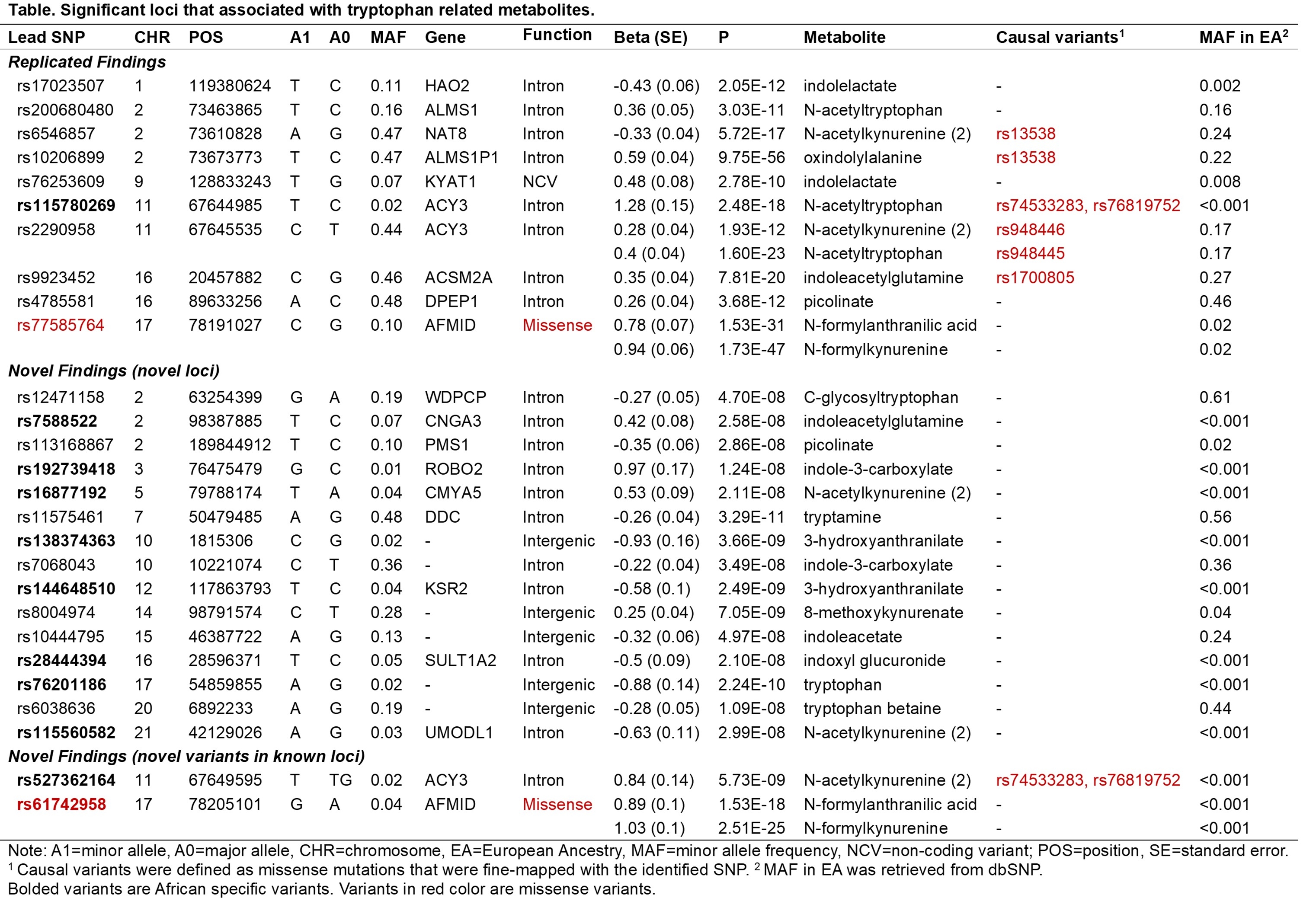
Results: Twenty-seven independent variants were identified for 17 tryptophan-related metabolites (Table). Seventeen, including 10 African specific, variants were novel. Of note, ACY3 intronic variant rs527362164 was associated with N-acetylkynurenine and was fine-mapped to two missense mutations, rs74533283 and rs76819752, in ACY3. The three variants were African specific (minor allele frequency [MAF]>1%) and missing in other populations (MAF<0.1%). Interestingly, the two missense variants were also the causal variants of a previously reported African-specific intronic variant, rs115780269, associated with plasma N-acetyltryptophan. Similarly, AFMID missense variant rs61742958 was significantly associated with N-formylanthranilic acid and N-formylkynurenine. This variant was also African specific and fine-mapped as the causal variant of a previously identified intronic variant, rs114080902, for plasma N-formylanthranilic. We also replicated 12 associations for 10 independent variants in well-established genes involved in tryptophan metabolism.
Conclusion: We identified 17 novel variants for tryptophan metabolism in African American CKD patients. Ten variants, including missense variants in AFMID and ACY3 genes, were African specific. Our findings underscore the importance of genetic studies in population subgroups to reveal novel mechanisms of tryptophan metabolism.
Methods: Thirty-two tryptophan-related metabolites were profiled from baseline 24-hour urine samples. Genome-wide array data were imputed to the Trans-Omics for Precision Medicine reference panel. After stringent quality control, GWAS of each metabolite was conducted, adjusting for age, sex, and 10 genetic principal components. Variants with P < 5×10-8 were considered significant. Conditional association analysis using GCTA-cojo, followed by fine-mapping using SuSiE, was performed on the 1000 kb region around a significant variant to identify independent signals and causal variants.
Results: Twenty-seven independent variants were identified for 17 tryptophan-related metabolites (Table). Seventeen, including 10 African specific, variants were novel. Of note, ACY3 intronic variant rs527362164 was associated with N-acetylkynurenine and was fine-mapped to two missense mutations, rs74533283 and rs76819752, in ACY3. The three variants were African specific (minor allele frequency [MAF]>1%) and missing in other populations (MAF<0.1%). Interestingly, the two missense variants were also the causal variants of a previously reported African-specific intronic variant, rs115780269, associated with plasma N-acetyltryptophan. Similarly, AFMID missense variant rs61742958 was significantly associated with N-formylanthranilic acid and N-formylkynurenine. This variant was also African specific and fine-mapped as the causal variant of a previously identified intronic variant, rs114080902, for plasma N-formylanthranilic. We also replicated 12 associations for 10 independent variants in well-established genes involved in tryptophan metabolism.
Conclusion: We identified 17 novel variants for tryptophan metabolism in African American CKD patients. Ten variants, including missense variants in AFMID and ACY3 genes, were African specific. Our findings underscore the importance of genetic studies in population subgroups to reveal novel mechanisms of tryptophan metabolism.
More abstracts on this topic:
Ambulatory Blood Pressure Variability, Progression of Kidney Disease, and Cardiovascular Outcomes in the Chronic Renal Insufficiency Cohort
Byfield Rushelle, Cohen Debbie, Townsend Raymond, Zhang Rachel, Hossain Alavi, Shimbo Daichi, Cohen Jordana
Association of Eicosanoid Metabolites with Body Mass IndexChitsazan Mandana, Ho Jennifer, Parekh Juhi, Lau Emily, Alotaibi Mona, Yu Bing, Allen Norrina, Allison Matthew, Jain Mohit, Cheng Susan