Final ID: Mo4063
Integrated Radiogenomics Improves Classification of Genetic Hypertrophic Cardiomyopathy
Abstract Body (Do not enter title and authors here): Background
The prognosis of hypertrophic cardiomyopathy (HCM) varies by several factors, including imaging characteristics and genotype, reflecting underlying pathology. Traditional machine learning models typically require large datasets, which are challenging to obtain in rare disease research. Bayesian Networks (BN) excel with limited data by incorporating domain knowledge and handling imaging uncertainty.
Research Question
Can integrated radiogenomics improve the classification of genetic variants in HCM compared to models using clinical or radiomic features alone?
Aim
To establish a novel BN that evaluates the ability of radiogenomics to improve disease classification by integrating imaging derived radiomic features with clinical genotyping data.
Methods
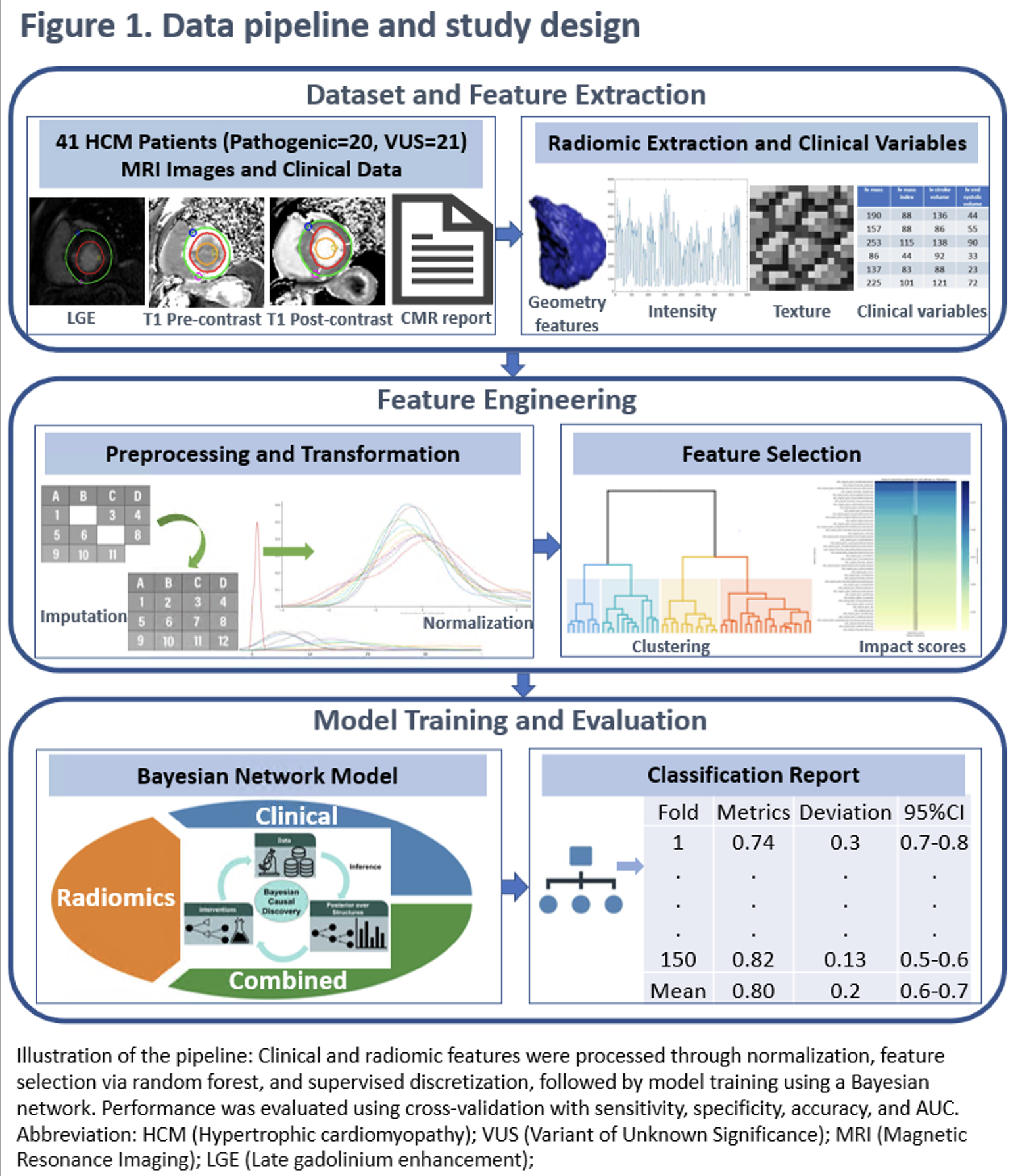
We used a BN to analyze 41 HCM patients who had both clinical cardiovascular magnetic resonance (CMR, 3T, Siemens Health Systems, Germany) and genotyping (Ambry Genetics, Aliso Viejo, CA; Lapcorp Invitae, San Francisco, CA), identifying 20 Pathogenic/likely pathogenic (P/LP) and 21 VUS between 2018-2024. CMR included pre- and post-contrast T1 mapping and late gadolinium enhancement (LGE) sequences. Clinical and CMR variables included cardiac structure and function, body size, blood pressure, heart rate, age, and sex. Clinical variables and radiomic features from T1 and LGE were extracted, normalized, and reduced using mutual information and random forest feature importance. A BN with domain-informed priors trained using 5-fold cross-validation (CV) (30 repeats) classified P/LP vs. VUS, reporting sensitivity, specificity, accuracy and area under the curve (AUC) (Figure 1).
Results
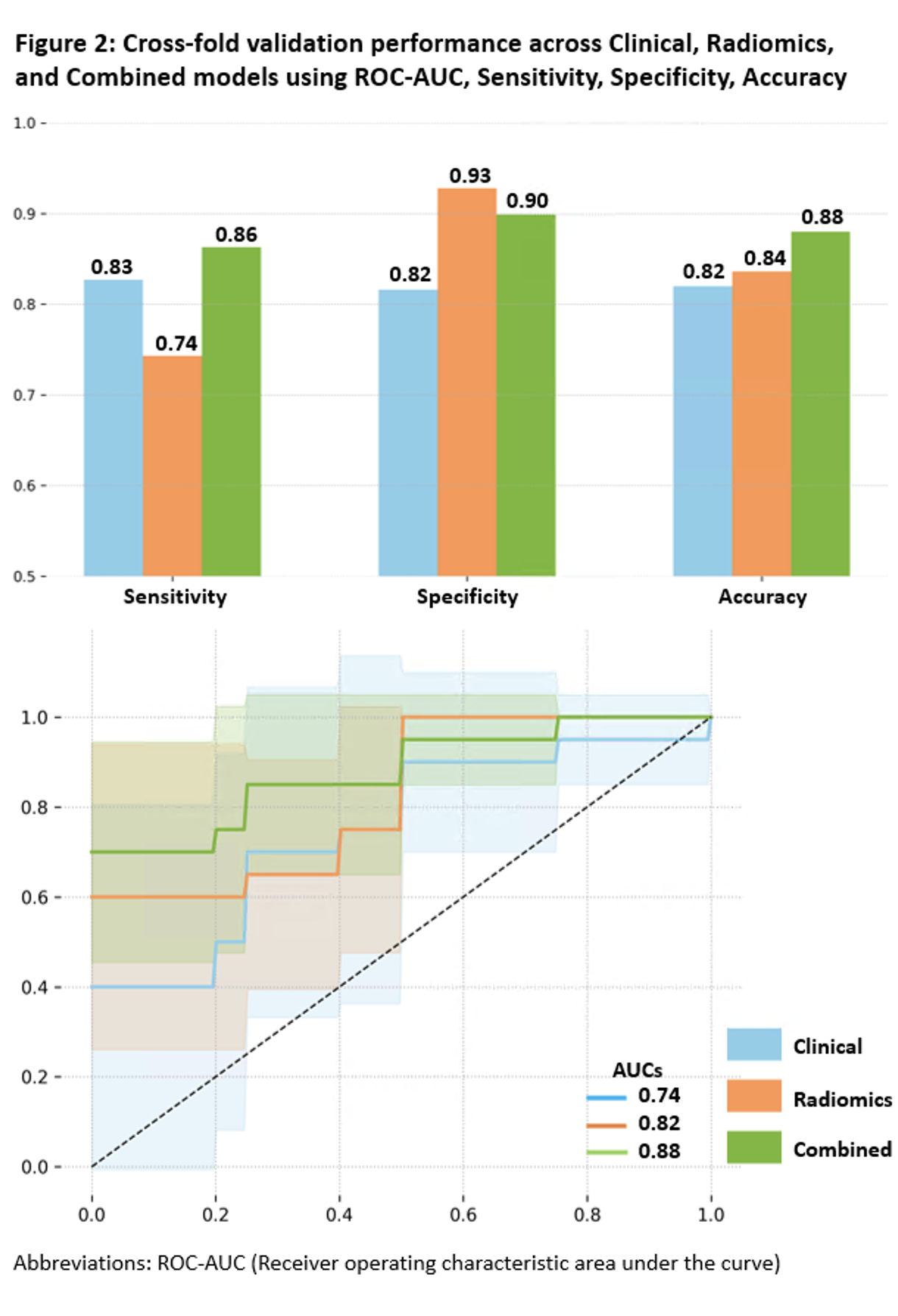
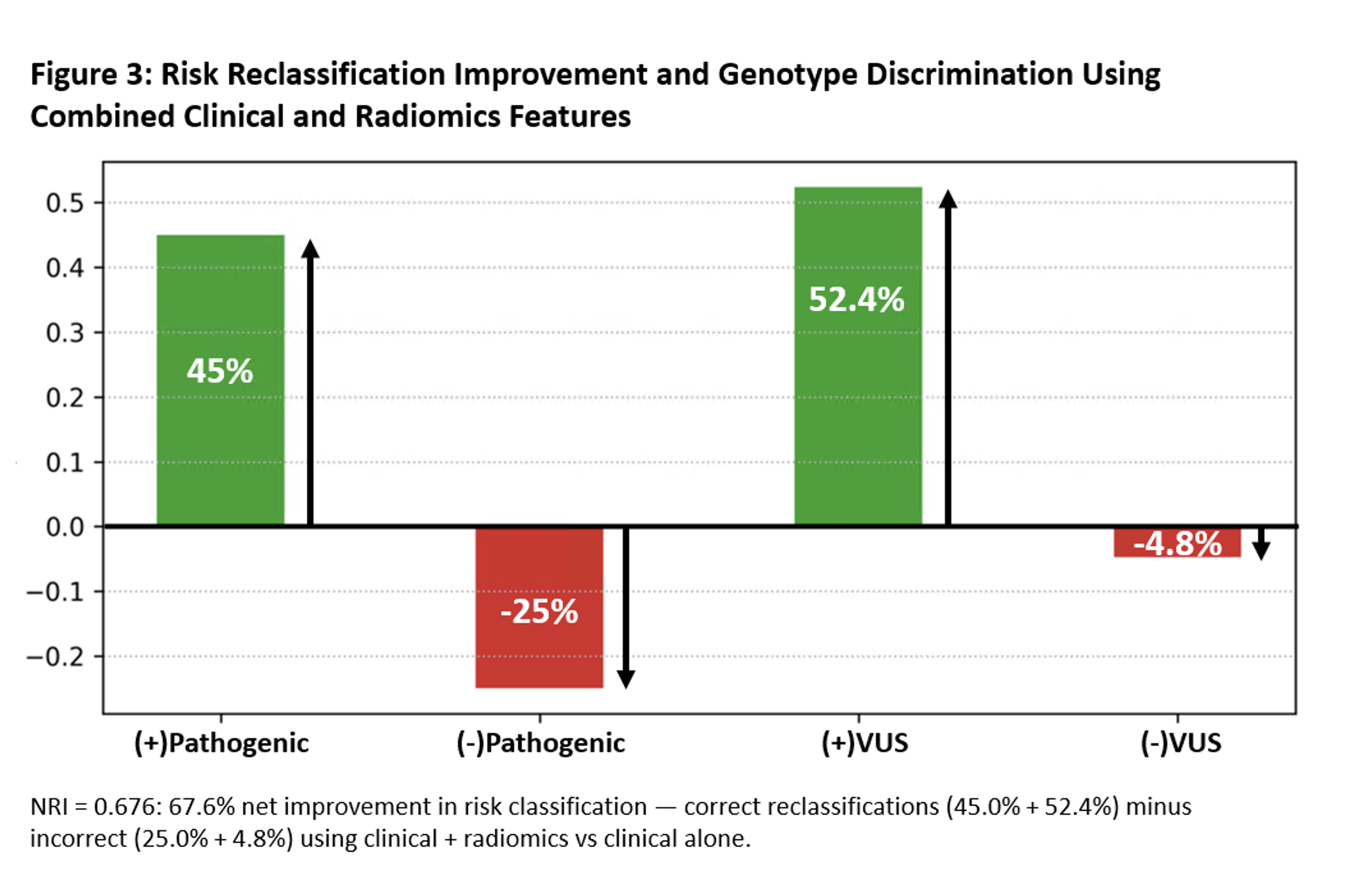
The radiomics model for classifying P/LP vs. VUS in HCM achieved a mean CV accuracy of 0.84 and an AUC of 0.82, outperforming the clinical model (0.82 accuracy, 0.74 AUC). The combined clinical+radiomics model improved further (0.88 accuracy, 0.88 AUC) (Figure 2), with a net reclassification index of 0.68, demonstrating the strength of integrated modeling (Figure 3).
Conclusions
Integrated radiogenomics using BNs shows promise in classifying HCM variants with limited data, improving performance above clinical models. External validation will further refine these findings.
The prognosis of hypertrophic cardiomyopathy (HCM) varies by several factors, including imaging characteristics and genotype, reflecting underlying pathology. Traditional machine learning models typically require large datasets, which are challenging to obtain in rare disease research. Bayesian Networks (BN) excel with limited data by incorporating domain knowledge and handling imaging uncertainty.
Research Question
Can integrated radiogenomics improve the classification of genetic variants in HCM compared to models using clinical or radiomic features alone?
Aim
To establish a novel BN that evaluates the ability of radiogenomics to improve disease classification by integrating imaging derived radiomic features with clinical genotyping data.
Methods
We used a BN to analyze 41 HCM patients who had both clinical cardiovascular magnetic resonance (CMR, 3T, Siemens Health Systems, Germany) and genotyping (Ambry Genetics, Aliso Viejo, CA; Lapcorp Invitae, San Francisco, CA), identifying 20 Pathogenic/likely pathogenic (P/LP) and 21 VUS between 2018-2024. CMR included pre- and post-contrast T1 mapping and late gadolinium enhancement (LGE) sequences. Clinical and CMR variables included cardiac structure and function, body size, blood pressure, heart rate, age, and sex. Clinical variables and radiomic features from T1 and LGE were extracted, normalized, and reduced using mutual information and random forest feature importance. A BN with domain-informed priors trained using 5-fold cross-validation (CV) (30 repeats) classified P/LP vs. VUS, reporting sensitivity, specificity, accuracy and area under the curve (AUC) (Figure 1).
Results
The radiomics model for classifying P/LP vs. VUS in HCM achieved a mean CV accuracy of 0.84 and an AUC of 0.82, outperforming the clinical model (0.82 accuracy, 0.74 AUC). The combined clinical+radiomics model improved further (0.88 accuracy, 0.88 AUC) (Figure 2), with a net reclassification index of 0.68, demonstrating the strength of integrated modeling (Figure 3).
Conclusions
Integrated radiogenomics using BNs shows promise in classifying HCM variants with limited data, improving performance above clinical models. External validation will further refine these findings.
More abstracts on this topic:
Abnormal Calcium Regulation Leads to Pathological Cardiac Hypertrophy During Pregnancy in the GSNOR-Deficient Mouse Model of Preeclampsia
Dulce Raul, Balkan Wayne, Hare Joshua, Kulandavelu Shathiyah
A Novel Variant in GNB2 as a Cause of Sick Sinus SyndromeBulut Aybike, Karacan Mehmet, Saygili E. Alper, Pirli Dogukan, Aydin Eylul, Ozdemir Ozkan, Balci Nermin, Alanay Yasemin, Bilguvar Kaya, Akgun Dogan Ozlem