Final ID: 4146799
Medical Applications of Artificial Intelligence: Coronary Artery Segmentation, Lesion Identification and Measurement in X-Ray Angiography
Abstract Body (Do not enter title and authors here): Background
Coronary artery segmentation, Lesion Identification and Measurement (CASLIM) on XRA images on X-ray angiography (XRA) are performed by cardiologists.
Aims
The study, CASLIM aims to develop an end-to-end deep learning framework with integrated conventional image processing methods that accurately performs those tasks on XRA images.
Methods
In depth analysis and comparisons of the deep learning models UNet, UNet+, Attention UNet, Trans UNet, SwinUNet, and UNet3+ comprising of the training curve and validation curve plotted against the epochs were carried out. The testing scores are obtained from the results of trained models on around 80 full size images. The data is presented for validity of the UNet3+ model as compared to the others.
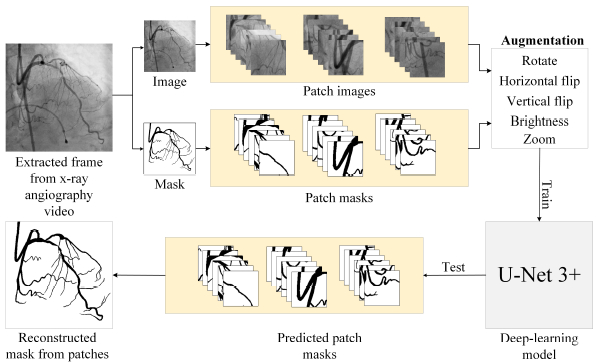
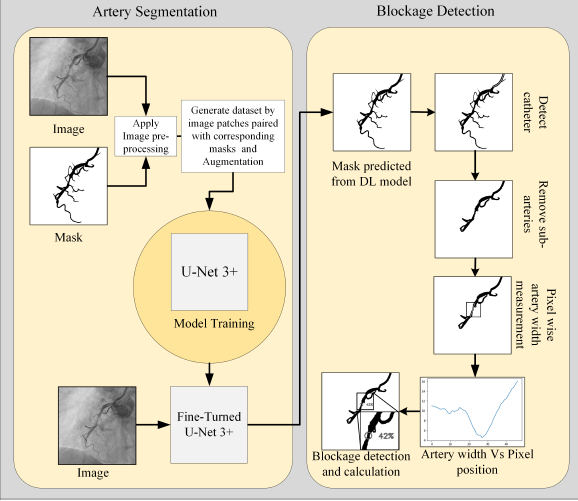
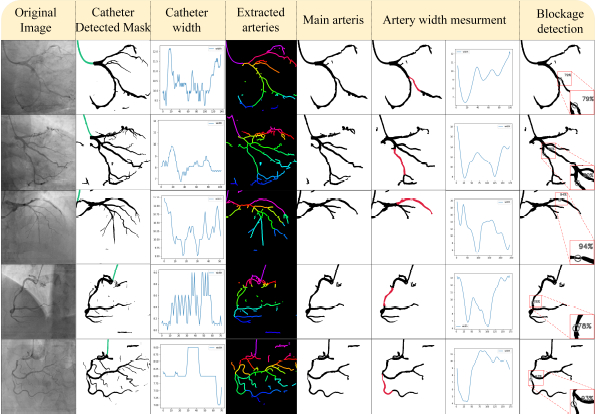
The performance of the proposed CASLIM analysis starts with the training results of the UNet3+ and other models for the coronary pixel segmentation. For UNet3+, patch sizes of 128*128 and 244*244 are studied to define the effect of patch sizes on performance. The model is trained on XRA images for accurate segmentation. (Image-1) Multiple image processing techniques are developed for lesion assessment. A dataset for training, validation and testing of the model is prepared by manual pixel annotation. Image processing algorithms are innovated to develop algorithms for catheter detection, its width measurement; subtract small arteries; thinning and pixel width measurement on binary mask images of arteries; detect, locate and quantify lesions. (Images-2,3) Experiments evaluated this method by comparing the lesion measurement results with the manually performed ones.
Results
The dice score, 0.989; Structural Similarity Index Measurement (SSIM), 0.888; SSIM loss, 0.809 and Intersection Over Union, 0.979 of UNet3+ show its excellent performance and proximity to the ground truth in segmentation; overcoming the issues of intensity and position variations, noise and overlapping structures. The numerical testing performance report shows that the UNet3+ with a patch size of 128*128 performs the best. The lesion measurement by CASLIM shows a mean square error (MSE) value- 28.66 and an R squared (R2) value- 0.81 as compared to the manual process. The MSE- 69.91 and R2- 0.99 are obtained with the proposed method for lesion localization as compared to the manual process.
Conclusion
The UNet3+ model has exceptional accuracy for precise segmentation. The CASLIM presents a promising solution for automated coronary lesion assessment accurately.
Coronary artery segmentation, Lesion Identification and Measurement (CASLIM) on XRA images on X-ray angiography (XRA) are performed by cardiologists.
Aims
The study, CASLIM aims to develop an end-to-end deep learning framework with integrated conventional image processing methods that accurately performs those tasks on XRA images.
Methods
In depth analysis and comparisons of the deep learning models UNet, UNet+, Attention UNet, Trans UNet, SwinUNet, and UNet3+ comprising of the training curve and validation curve plotted against the epochs were carried out. The testing scores are obtained from the results of trained models on around 80 full size images. The data is presented for validity of the UNet3+ model as compared to the others.
The performance of the proposed CASLIM analysis starts with the training results of the UNet3+ and other models for the coronary pixel segmentation. For UNet3+, patch sizes of 128*128 and 244*244 are studied to define the effect of patch sizes on performance. The model is trained on XRA images for accurate segmentation. (Image-1) Multiple image processing techniques are developed for lesion assessment. A dataset for training, validation and testing of the model is prepared by manual pixel annotation. Image processing algorithms are innovated to develop algorithms for catheter detection, its width measurement; subtract small arteries; thinning and pixel width measurement on binary mask images of arteries; detect, locate and quantify lesions. (Images-2,3) Experiments evaluated this method by comparing the lesion measurement results with the manually performed ones.
Results
The dice score, 0.989; Structural Similarity Index Measurement (SSIM), 0.888; SSIM loss, 0.809 and Intersection Over Union, 0.979 of UNet3+ show its excellent performance and proximity to the ground truth in segmentation; overcoming the issues of intensity and position variations, noise and overlapping structures. The numerical testing performance report shows that the UNet3+ with a patch size of 128*128 performs the best. The lesion measurement by CASLIM shows a mean square error (MSE) value- 28.66 and an R squared (R2) value- 0.81 as compared to the manual process. The MSE- 69.91 and R2- 0.99 are obtained with the proposed method for lesion localization as compared to the manual process.
Conclusion
The UNet3+ model has exceptional accuracy for precise segmentation. The CASLIM presents a promising solution for automated coronary lesion assessment accurately.
More abstracts on this topic:
A Machine Learning Approach to Predict Percutaneous Coronary Intervention in Patients with Critical Illness and Signs of Myocardial Injury
Mueller Joshua, Stepanova Daria, Chidambaram Vignesh, Nakarmi Ukash, Al'aref Subhi
A large-scale multi-view deep learning-based assessment of left ventricular ejection fraction in echocardiographyJing Linyuan, Metser Gil, Mawson Thomas, Tat Emily, Jiang Nona, Duffy Eamon, Hahn Rebecca, Homma Shunichi, Haggerty Christopher, Poterucha Timothy, Elias Pierre, Long Aaron, Vanmaanen David, Rocha Daniel, Hartzel Dustin, Kelsey Christopher, Ruhl Jeffrey, Beecy Ashley, Elnabawi Youssef