Final ID: MDP791
Random survival forests identify myocardial gene signatures associated with survival in heart failure with preserved ejection fraction
Abstract Body (Do not enter title and authors here): Background: Heart failure with preserved ejection fraction (HFpEF) continues to be poorly understood at the molecular level. While previous studies have identified gene expression signatures unique to HFpEF, genes associated with clinical decompensation have not been determined. Here, we performed exploratory analysis of myocardial RNA-seq data to identify genes associated with event-free survival in HFpEF.
Methods: We analyzed previously published RNA sequencing data of right ventricular septal endomyocardial biopsies from HFpEF patients (n=41) with paired clinical, echocardiographic, and outcome data (including mortality and heart failure hospitalizations). We constructed random survival forests with forward stepwise regularization (the “variable hunting” method) to determine genes associated with time to first event using a combined end-point of heart failure hospitalization or all-cause death. Selection of candidate forest variables was tested with both random and weighted sampling methods.
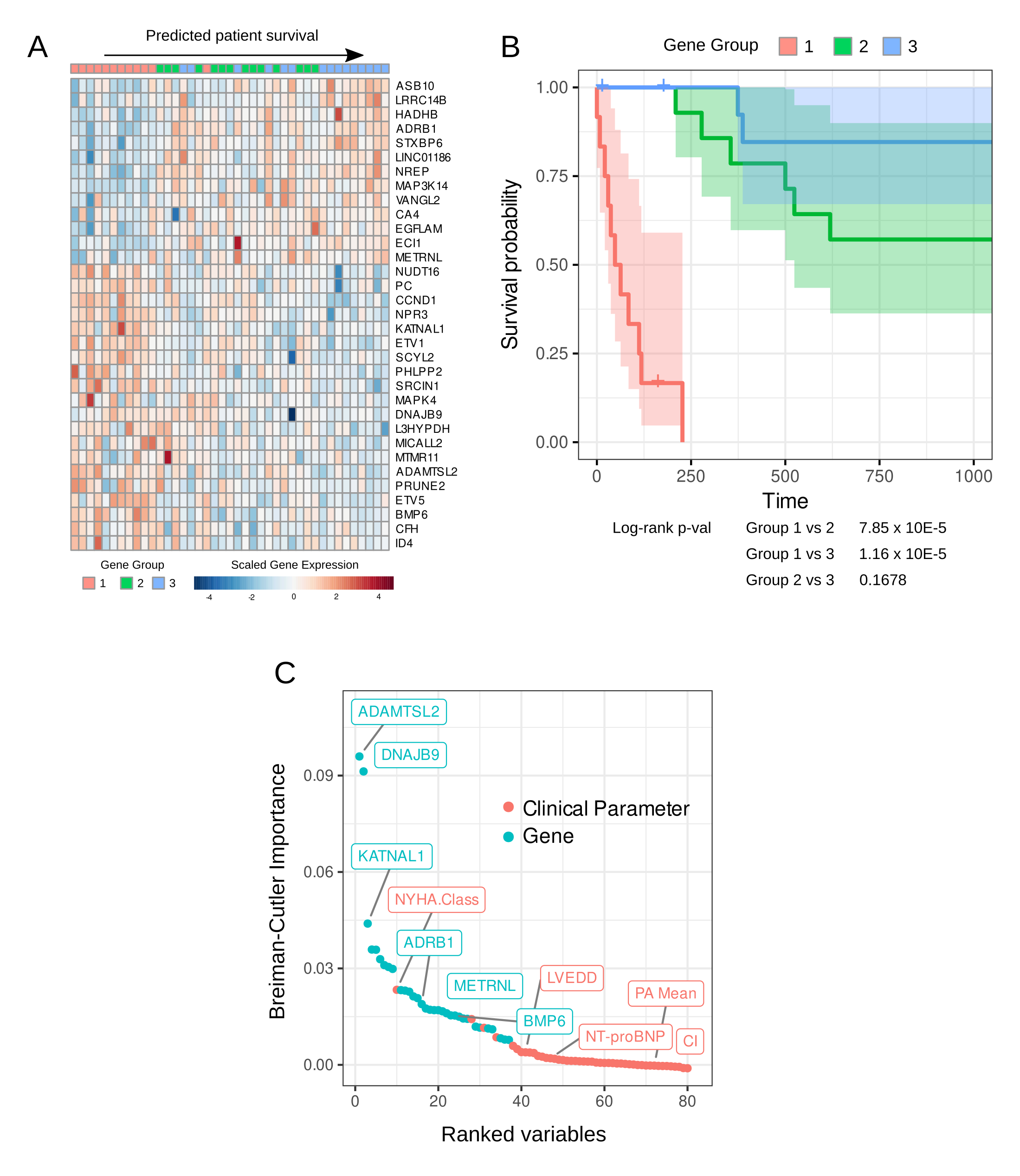
Results: We identified 33 genes that are predictive of survival in HFpEF (Figure 1A). This set includes genes previously implicated in heart failure, including ADAMTSL2, ADRB1, BMP6, and METRNL. Patients with HFpEF were categorized into 3 groups (Gene Group) based on their expression of this survival gene signature. Gene Group 1 had the worst survival compared to Gene Groups 2 and 3 (log rank p values 7.85e-5 for Gene Group 1 vs 2, 1.16e-5 for Gene Group 1 vs 3, adjusted for age, sex, and renal function, Figure 1B). Survival was not significantly different between Gene Groups 2 and 3 (adjusted log-rank p value 0.17). Survival forests constructed using these genes outperform those constructed from clinical and hemodynamic parameters alone (out-of-bag C-index 0.894 vs 0.545). Moreover, in survival forests constructed from both gene data and hemodynamic measurements, individual survival genes consistently showed higher variable importance than clinical/hemodynamic parameters (Figure 1C).
Conclusions: Random survival forests identify myocardial gene signatures that may better model HFpEF prognosis than clinical measurements. Further studies are warranted to validate findings in independent cohorts.
Methods: We analyzed previously published RNA sequencing data of right ventricular septal endomyocardial biopsies from HFpEF patients (n=41) with paired clinical, echocardiographic, and outcome data (including mortality and heart failure hospitalizations). We constructed random survival forests with forward stepwise regularization (the “variable hunting” method) to determine genes associated with time to first event using a combined end-point of heart failure hospitalization or all-cause death. Selection of candidate forest variables was tested with both random and weighted sampling methods.
Results: We identified 33 genes that are predictive of survival in HFpEF (Figure 1A). This set includes genes previously implicated in heart failure, including ADAMTSL2, ADRB1, BMP6, and METRNL. Patients with HFpEF were categorized into 3 groups (Gene Group) based on their expression of this survival gene signature. Gene Group 1 had the worst survival compared to Gene Groups 2 and 3 (log rank p values 7.85e-5 for Gene Group 1 vs 2, 1.16e-5 for Gene Group 1 vs 3, adjusted for age, sex, and renal function, Figure 1B). Survival was not significantly different between Gene Groups 2 and 3 (adjusted log-rank p value 0.17). Survival forests constructed using these genes outperform those constructed from clinical and hemodynamic parameters alone (out-of-bag C-index 0.894 vs 0.545). Moreover, in survival forests constructed from both gene data and hemodynamic measurements, individual survival genes consistently showed higher variable importance than clinical/hemodynamic parameters (Figure 1C).
Conclusions: Random survival forests identify myocardial gene signatures that may better model HFpEF prognosis than clinical measurements. Further studies are warranted to validate findings in independent cohorts.
More abstracts on this topic:
Artificial Intelligence-Enhanced Electrocardiographic Phenotyping Unveils Novel Lethal Cardiovascular & Sudden Death Risk Signatures in Traditionally Low Risk Populations
Barker Joseph, Syan Jasjit, Jenkins Alex, Ribeiro Antonio, Annis Jeffrey, Camelo Lidyane, Oliveira Clara, Paixao Gabriela, Brant Luisa, Ribeiro Antonio, Ge Junbo, Fathieh Sina, Kramer Daniel, Waks Jonathan, Brittain Evan, Peters Nicholas, Figtree Gemma, Khattak Gul Rukh, Sau Arunashis, Ng Fu Siong, Pastika Libor, Birdi Aidan, Zeidaabadi Boroumand, Patlatzoglou Konstantinos, Liang Yixiu, Aggour Hesham, El-medany Ahmed
Artificial Intelligence Predicted Age using the Electrocardiogram Predicts Mortality in Adults with Congenital Heart Disease.Anjewierden Scott, Madhavan Malini, Attia Zachi, Lopez-jimenez Francisco, Friedman Paul, Egbe Alexander, Connolly Heidi, Asirvatham Samuel, Burchill Luke