Final ID: MDP790
Performance of a Protein Language Model for Variant Annotation in Cardiac Disease
Abstract Body (Do not enter title and authors here): Introduction: Genetic testing is a cornerstone in the assessment of many cardiac diseases. However, variants are frequently classified as Variants of Unknown Significance (VUS), limiting the utility of testing.
Recently, the DeepMind group (Google, USA) developed AlphaMissense, a unique Artificial Intelligence (AI) based model, based on language model principles for the prediction of missense variant pathogenicity.
Objective: To report on the performance of AlphaMissense, accessed by VarCardio, an open web-based variant annotation engine, in a real-world cardiovascular genetics center.
Methods: All genetic variants from an inherited arrhythmia program were examined using AlphaMissense via VarCard.io and compared to the ClinVar variant classification system, as well as another variant classification platform (Franklin by Genoox). The mutation reclassification rate and genotype-phenotype concordance were examined for all variants in the study.
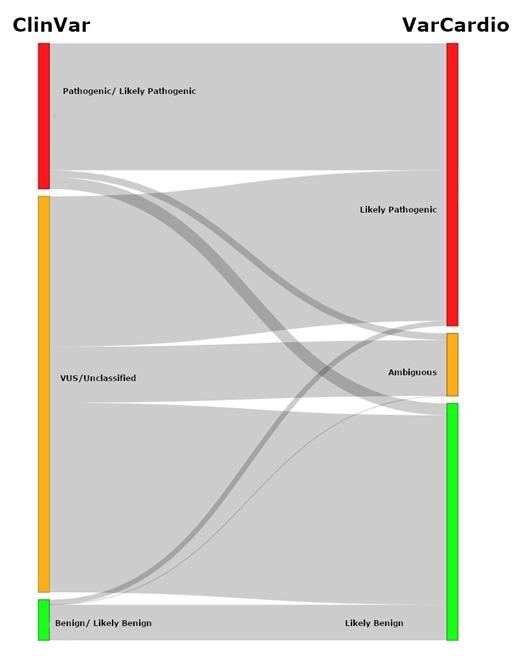
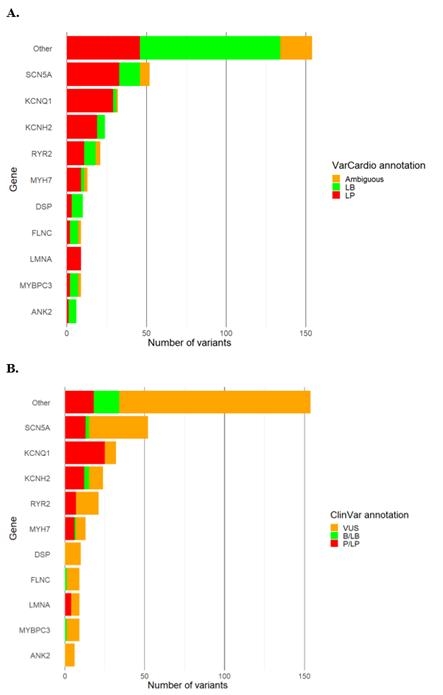
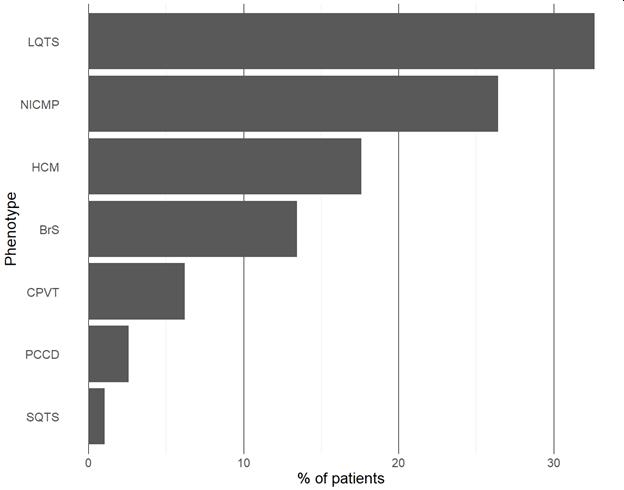
Results: We included 266 patients with heritable cardiac diseases, harboring 339 missense variants. Of those, 230 (67.8%) were classified by ClinVar as either VUS or non-classified. Using VarCard.io, 198 VUSs (86.1%, CI 80.9-90.3%) were reclassified to either Likely Pathgenic (LP) or Likely benign (LB). The reclassification rate was significantly higher for VarCard.io than for Franklin (86.1% vs 34.8%, p<0.001).
Genotype-Phenotype concordance was highly aligned using VarCard.io predictions, at 95.9% (CI 92.8-97.9%) concordance rate. For 109 variants classified as Pathogenic, LP, Benign or LB by ClinVar, concordance with VarCard.io was high (90.5%).
Conclusion: AlphaMissense, accessed via VarCard.io, may be a highly efficient tool for cardiac genetic variant interpretation. The engine's notable performance in assessing variants that are classified as VUS in ClinVar, demonstrates its potential to enhance cardiac genetic testing.
Recently, the DeepMind group (Google, USA) developed AlphaMissense, a unique Artificial Intelligence (AI) based model, based on language model principles for the prediction of missense variant pathogenicity.
Objective: To report on the performance of AlphaMissense, accessed by VarCardio, an open web-based variant annotation engine, in a real-world cardiovascular genetics center.
Methods: All genetic variants from an inherited arrhythmia program were examined using AlphaMissense via VarCard.io and compared to the ClinVar variant classification system, as well as another variant classification platform (Franklin by Genoox). The mutation reclassification rate and genotype-phenotype concordance were examined for all variants in the study.
Results: We included 266 patients with heritable cardiac diseases, harboring 339 missense variants. Of those, 230 (67.8%) were classified by ClinVar as either VUS or non-classified. Using VarCard.io, 198 VUSs (86.1%, CI 80.9-90.3%) were reclassified to either Likely Pathgenic (LP) or Likely benign (LB). The reclassification rate was significantly higher for VarCard.io than for Franklin (86.1% vs 34.8%, p<0.001).
Genotype-Phenotype concordance was highly aligned using VarCard.io predictions, at 95.9% (CI 92.8-97.9%) concordance rate. For 109 variants classified as Pathogenic, LP, Benign or LB by ClinVar, concordance with VarCard.io was high (90.5%).
Conclusion: AlphaMissense, accessed via VarCard.io, may be a highly efficient tool for cardiac genetic variant interpretation. The engine's notable performance in assessing variants that are classified as VUS in ClinVar, demonstrates its potential to enhance cardiac genetic testing.
More abstracts on this topic:
A Multi-Population-First Approach Leveraging UK Biobank (UKBB) and All of Us (AoU) Datasets Reveals Higher Cardiomyopathy Variant Burden in Individuals with Myocarditis
Gurumoorthi Manasa, Khanji Mohammed, Munroe Patricia, Petersen Steffen, Landstrom Andrew, Chahal Anwar, Hesse Kerrick, Asatryan Babken, Shah Ravi, Sharaf Dabbagh Ghaith, Wolfe Rachel, Shyam Sundar Vijay, Mohiddin Saidi, Aung Nay
A Multi-Population-First Approach Leveraging UK Biobank (UKBB) and All of Us (AoU) Datasets Reveals Higher Cardiomyopathy Variant Burden in Individuals with MyocarditisGurumoorthi Manasa, Khanji Mohammed, Munroe Patricia, Petersen Steffen, Landstrom Andrew, Chahal Anwar, Hesse Kerrick, Asatryan Babken, Shah Ravi, Sharaf Dabbagh Ghaith, Wolfe Rachel, Shyam Sundar Vijay, Mohiddin Saidi, Aung Nay